CellML.org - CellML Specification : Draft - 2 March 2001
CellML Specification Overview Entire Specification Introduction Fundamentals Model Structure Mathematics Units Grouping Reactions Metadata Framework |
This Version: Status of this documentThis document is a preliminary version of the specification for version 1.0 of CellMLTM. It is distributed for discussion purposes only, and should not be used as a reference. The definitive version of the CellML specification can always be found on the CellML website: http://www.cellml.org/public/specification/index.html A message will be posted to the cellml-announce mailing list when a full candidate specification is available. You can join the cellml-announce mailing list at: http://www.cellml.org/mailman/listinfo. Feedback on this document should be sent to the cellml-discussion mailing list. You can join this mailing list at: http://www.cellml.org/mailman/listinfo. Note that members of the cellml-discussion mailing list will receive all posts to cellml-announce, so it is not necessary to join both lists. If you wish to provide feedback, but do not want to join the cellml-discussion mailing list, you can e-mail your comments to mailto:info@cellml.org. All sections of this specification are subject to change. The following items are currently under review, and will almost certainly change:
In addition, we intend to add the following content to the specification before the full candidate specification is released:
Contents
1 Introduction1.1 Introduction to CellMLThis document formally specifies CellMLTM, an XML-based language for describing and exchanging a wide range of mathematical models of cells and subcellular processes. CellML is being developed by scientists in the Bioengineering Research Group at the University of Auckland and at Physiome Sciences, Inc. The development of CellML is guided by an advisory board drawn from many different areas of biological modelling (see the project team page on the CellML website for more information). CellML is being developed as an open standard, and all interested parties are encouraged to send feedback to info@cellml.org, or to the cellml-discussion mailing list. 1.1.1 Purpose and scope of CellMLCellML is intended to support the definition of any type of model of a cell or subcellular process. Therefore, it uses a very general structure. CellML is also intended to facilitate the re-use of models and parts of models. It accomplishes this by using a component-based architecture. Models are split into logical sub-parts called components. These components are then connected together to form a model. The scope of CellML is specifically limited to the definition of model structure. All other types of information that modellers need or want to include in a model document are incorporated using other languages. For instance, mathematics is included in CellML documents using MathML. Metadata is included as RDF, using the Dublin Core's schema wherever possible. 1.1.2 What is XML?The CellML language is defined in terms of a meta-language called XML, which stands for eXtensible Markup Language. XML is a standard published by the World Wide Web Consortium, the organisation responsible for defining many internet-related standards, most notably HTML. XML is essentially a means of adding structure to text documents, allowing machines to unambiguously associate text or binary data with a particular component in a document's data model. XML is an appropriate medium for CellML because it is both human and machine readable. A model author can create a CellML document with a text editor or with any piece of CellML-compliant software. XML is a well-defined and widely used specification, and many free software utilities and libraries for the processing of XML already exist, simplifying the development of CellML software. XML has also been designed to be usable over the internet, making CellML suitable for the interchange of models between software and databases at different physical locations. A quick introduction to XML is available in the examples section of the CellML website. 1.1.3 Definition of "model"A model is an idealized representation of the rules that govern the behaviour of a system. CellML supports both quantitative and qualitative models. Quantitative models represent these rules using mathematics. Qualitative models represent the relationships between objects in the system, without attempting to define mathematics to represent the behaviour of the objects. The CellML specification covers three kinds of models: complete, incomplete, and partial. A complete quantitative model is one that can be simulated (i.e., the mathematical equations contained in the model can be solved). A complete qualitative model is one in which all objects of interest in a system are represented. An incomplete model is a work in progress. For instance, an incomplete quantitative model might not contain all of the equations necessary to simulate the behaviour of the system. A partial model is a description of one aspect of the system. Within that portion of the system, the description is complete. However, it still might not be possible to run a simulation of the model. A valid CellML document may describe a complete, incomplete, or partial model. A valid CellML model must be complete. This specification does not attempt to limit the behaviour of processing software when confronted with invalid documents or models. It is recommended that software report errors to the modeller (at the very least). 1.2 Structure of the CellML Specification1.2.1 Sections of the CellML specificationThe CellML specification is divided into several sections, each of which discusses a particular aspect of CellML:
A valid CellML model can be created using nothing beyond the material covered in the fundamentals, basic model structure, mathematics, and units sections of the specification. The concepts in the remaining sections of the specification allow modellers to build more meaningful models. Each section of the specification is further divided into five subsections:
1.2.2 Levels of CellML conformanceThe rules in the CellML specification can be split into two groups: rules that define the syntax of a CellML document and rules that determine how software processing that document should behave. In the subsequent sections of the specification, the first set of rules are included in subsections titled Rules for CellML Documents, and the second set is in subsections titled Rules for Processor Behaviour. The rules can also be split into two groups, each representing different levels of conformance to the specification. The majority of the rules in the CellML specification are part of the first level of conformance. Rules that are part of the second level of conformance are indicated as such by the inclusion of the phrase "Level Two" after the rule statement. The meaning of these conformance levels for documents and processing software is discussed below. The levels of conformance to the CellML specification should not be confused with the features defined in different versions of the specification. As CellML is developed further, future versions of the CellML specification will add new elements to the language, which may add document and behaviour rules that affect both levels of conformance. Features that are expected to be added to CellML are documented in the Future Directions part of the CellML website. CellML conformance level one
The first level of CellML conformance is composed of the majority of rules in the CellML specification. Level one document rules generally specify how the different XML elements and attributes that make up the CellML vocabulary may be combined. A typical level one rule for a document is "Both the A CellML Document is conformant to level one of the CellML specification if it complies with all level one rules for documents in the CellML specification. A CellML processor is conformant to level one of the CellML specification if it can validate CellML documents against all level one rules for documents in the CellML specification and it follows all appropriate level one rules for processor behaviour in the CellML specification when interpreting CellML documents. The appropriate rules are those that relate to the intended use of the software (i.e., software that only renders the model need not address the scope of units definitions). CellML conformance level twoThe second level of CellML conformance rule is composed of all of the rules from level one plus additional rules that are marked as belonging to level two in the specification. Currently, there are no level two document rules. Level two processor rules generally specify complex interactions between objects defined in different parts of a CellML document. A typical example is the requirement that all mathematics within a model be self-consistent. A CellML document is conformant to level two of the CellML specification if it complies with all level one and level two rules for documents in the CellML specification. A CellML processor is conformant to level two of the CellML specification if it can validate CellML documents against all level one and level two rules for documents in the CellML specification and it follows all appropriate level one and level two rules for processor behaviour in the CellML specification when interpreting CellML documents. The appropriate rules are those that relate to the intended use of the software (i.e., software that only renders the model need not address the consistency of mathematics). 2 Fundamentals2.1 IntroductionThe fundamentals section of the CellML specification introduces some concepts that are used throughout the entire language, and defines rules that are referenced in all or many of the other parts of the specification. These include the definition of names in CellML and recommended practice for the use of namespaces in CellML. 2.2 Basic Structure2.2.1 Definition of a valid CellML identifier
The most common use of a CellML identifier is the
The generation of computer code for running simulations is one of the target applications for CellML. The value of an object's The XML specification is based on the Unicode standard, which defines a scheme for 16 bit character encoding. Thus it is possible to include, for instance, Japanese characters in a valid XML document. In the interests of making the code generation process as convenient as possible for those using mainstream programming languages, CellML identifiers are subject to the following constraints:
Convenient code generation is also the reason why colons, periods, and hyphens may not appear in CellML identifier. CellML identifiers are case sensitive: a variable with an identifier of The specification of a valid CellML identifier is identical to the definition of a valid object name in SBML. This should simplify the process of translating model definitions between the two languages. 2.2.2 Namespaces in CellMLNamespaces in XML is a companion specification to the main XML specification. It provides a facility for associating the elements and/or attributes in all or part of a document with a particular schema, as indicated by a Uniform Resource Identifier (URI). The key aspect of the URI is that it is unique. The value of the URI need not have anything to do with the XML document that uses it, although typically it would be a good location for the XML Schema or DTD that defines the rules for the document type. The URI may be mapped to a prefix, which may then be used in front of element and attribute names, separated by a colon. If not mapped to a prefix, the URI sets the default schema for the current element and all of its children. The CellML specification defines a small number of elements and attributes and a namespace with which they must be associated. Associating CellML elements and attributes with the CellML namespace allows them to be differentiated from elements and attributes from other vocabularies with which CellML syntax might be combined in a CellML document. For instance, CellML makes use of the MathML vocabulary for the definition of equations, and all MathML elements must be placed in the MathML namespace in order for CellML processing software to recognise those elements. Applications that store their own proprietary data within a CellML document must define their own namespaces, and associate their own elements and attributes with those namespaces, as discussed in Section 2.2.3. The scope of CellML is specifically limited to the definition of model structure. The CellML namespace includes all elements and attributes that define the structure of a model. All other information that may be included in a CellML document, such as mathematics and metadata, is included using other namespaces. Metadata is placed in a variety of namespaces, as described in Section 8. The MathML namespace is given special importance, because content in this namespace is considered as fundamental as content in the CellML namespace. An empty CellML element may not contain content in either the CellML or MathML namespace, although it may contain content in other namespaces, including the metadata namespaces defined in this specification. Table 1 defines all of the namespaces used in the rules defined in this specification. The first three namespaces are in the cellml.org domain, and are associated with the core model structure elements, some custom metadata elements, and an XML serialization of the Object Management Group's bibliographic query service (BQS) data model created for storing citations in CellML. The MathML and RDF namespaces are defined in standards administered by the World Wide Web Consortium. Finally, the Dublin Core and Dublin Core Qualifiers namespaces reference standards for metadata specification administered by the Dublin Core organisation.
Table 1 The CellML specification defines the expected behaviour of CellML processing software for XML elements and attributes that are in these namespaces. Applications may not place their own elements and attributes in these namespaces. See text for more details.
Table 1 also gives the recommended prefix to be mapped to each namespace declaration for use in CellML documents. It is recommended that when a CellML element such as 2.2.3 Extending CellML documentsCellML processing software may store information not covered by the CellML specification in a CellML document by defining its own elements and attributes and placing them in a namespace other than one of those defined in Table 1. (This specification only defines the content models of elements in the namespaces in Table 1 with respect to other elements in those namespaces.) Elements and attributes in extension namespaces may appear anywhere in a CellML document, as long as the result is well-formed XML. Because the CellML specification is only concerned with content in the CellML or MathML namespaces, elements in extension namespaces may even appear inside elements defined by the CellML specification to be empty. It is hoped that CellML processing applications will respect the extension elements and attributes of other applications. If a model is created in application A, which adds its own extension elements, and is subsequently edited in application B, it would be polite if application B included application A's extension elements in its output, even if these extension elements are now invalid. Applications will need to validate their own extension data if a CellML document is read in from a non-trusted location. The namespace extension mechanism provides a convenient way to associate a small amount of application-specific information with a model defined in CellML. However, it is recommended that applications needing to store large amounts of information, such as rendering or simulation information, do so in a separate document. This will make CellML documents easier to exchange, and will prevent the loss of application-specific information if the model is read into another application. 2.3 Examples
Figure 1 contains some example CellML elements, each of which defines a Figure 1
XML elements defining
Figure 2 contains portions of a typical CellML document that demonstrate the recommended use of namespaces. The root element sets the default namespace to the CellML namespace and also explicitly maps the CellML namespace to the Figure 2 A CellML fragment demonstrating the recommended use of namespaces in a CellML document. This fragment is taken from the simple electro-physiological model example on the CellML website.
Figure 3 demonstrates how software can embed its own information inside a valid CellML document using XML namespaces. The Figure 3 A CellML document demonstrating the use of XML namespaces to embed application specific data inside a CellML document. The extension namespace was invented for demonstration purposes only. 2.4 Rules for CellML Documents2.4.1 Valid CellML identifiers
2.4.2 Extension namespaces
2.4.3 Proper use of the CellML namespace
2.5 Rules for Processor Behaviour2.5.1 Treatment of CellML identifiers
2.5.2 Treatment of extension namespaces
3 Model Structure3.1 IntroductionAny model can be described as a network of connections between self-contained components. A component is a functional unit that corresponds to a physical compartment, event, or species or that is just a convenient modelling abstraction. Components contain variables and mathematical relationships that manipulate those variables. Connections contain mappings between the variables of connected components. 3.2 Basic Structure3.2.1 Definition of a model
A CellML model may be a complete, functional model; an incomplete model; or a partial model (as defined in Section 1.1.3). A model is declared with the Figure 4
The root element of an XML document containing a CellML model description is the
The
A
The 3.2.2 Definition of componentsConstructing a model from multiple components encourages the re-use of components. For instance, an electro-physiological model of a cell might be organised into components that represent various ion channels. All of the mathematics that describe the behaviour of the L-type calcium channel would be defined in a single component representing this particular ion channel. If a modeller wished to re-use the portion of the model representing the L-type calcium channel in another model, he or she would only need to copy this component.
A
A CellML
A
A
The definitions of two 3.2.3 Definition of variablesModels are usually developed to simulate the behaviour of a number of variables that have physiological significance. Each variable in the model belongs to a single component, which may contain equations or scripts that modify the value of that variable. The value of a variable may be passed through connections into other components. The variable must also be declared in these components, which can then use the value of the variable in their own equations and scripts but may not modify it.
The
A
When a variable is declared with either a
Whether or not a component may obtain the value of a variable in another component depends on the The components to which any given component may connect can be divided into three distinct classes. The set of all components encapsulated by the current component is referred to as the encapsulated set. If the current component is encapsulated, then the encapsulating component is referred to as the parent, and the set of all other components encapsulated by the same parent is referred to as the sibling set. If the current component is not encapsulated, then it has no parent and the sibling set consists of all other components in the model that are not encapsulated. The encapsulation hierarchy and its effects on variable mapping are described in Section 6. Eventually, it will be possible to specify the temporal and/or spatial variation of a variable's value using FieldML. The capability to include FieldML is still under development. At the present time, all variables must have singular values. 3.2.4 Definition of connectionsConnections provide the mechanism for mapping variables declared within one component to variables in another component, allowing information to be exchanged between the various components in the network. There will be many such mappings present in a network. The mapping of variables involves the transfer of a variable's value from one component to another. This transfer may involve a conversion to account for the units in which each component expects the variable's value to be defined. (More information on units conversion can be found in Section 5.)
The complete set of variable mappings between any two components constitutes a connection. Only one connection may be created between any two components in a model. Each connection contains a pair of component references, indicating the two components involved in the connection. The two component references each contain an ordered list of variable references. Each variable contained in the first ordered list is mapped to the corresponding variable contained in the second ordered list. Mapping depends only on the order of the
The
Two
The
The CellML example discussed in Section 3.3 demonstrates the definition of a 3.3 Examples
Figure 5 contains a portion of the CellML encoding of the Hodgkin-Huxley squid axon model published in 1952. The excerpt contains the definitions of the components corresponding to the membrane and the sodium channel, and the connection between the two components. Most of the complexity from the full model definition has been left out for conciseness and clarity. This example is only used to demonstrate the standard use of the Figure 5
A small portion of the CellML definition of the Hodgkin-Huxley squid axon model from 1952. This excerpt contains the definition of the components corresponding to the membrane and the sodium channel, and the connection between them. Much detail has been omitted, but this example clearly demonstrates the relationship between the
The
The second and third variables are
After the variable declarations, a
The
Finally, a 3.4 Rules for CellML Documents
The following are the rules for using the 3.4.1 The
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ampere | farad | katal | lux | pascal | tesla | |||||
| becquerel | gram | kelvin | meter | radian | volt | |||||
| candela | gray | kilogram | metre | second | watt | |||||
| celsius | henry | liter | mole | siemen | weber | |||||
| coulomb | hertz | litre | newton | sievert | ||||||
| dimensionless | joule | lumen | ohm | steradian |
Table 2 The dictionary of units keywords that CellML processing applications are expected to recognise. Base SI units are printed in bold text, derived SI units are printed in plain text, and additions to the standard units defined purely for the convenience of model authors are italicised.
This list is based on the ISO standard, including the year 2000 supplement. The American spellings of "meter" and "liter" are taken from the NIST Guide for the Use of the International System of UNITS (SI). The ISO standard defines the mathematical relationships between the derived SI units and the base SI units.
5.2.2 Definition of non-SI units
The CellML <units> elements in Figure 7 define the non-SI units in Table 2 (the italicised keywords) in terms of SI units. The format of the units element is formally specified in Section 5.2.3.
Figure 7
These <units> elements define the non-SI units included in the standard CellML units dictionary in terms of SI units. The definition of liter is exactly the same as the definition of litre, and is therefore omitted. Note that these definitions must not be used in CellML documents: model authors are forbidden from defining units with the same names as those in the standard dictionary in Table 2.
5.2.3 User defined units
CellML also provides a facility whereby new units can be defined in terms of the units provided in the dictionary. This functionality allows the definition of units which are expressed as a scaled version of other units (as is the case for most imperial units), the definition of units which are made up of the product of other units, and even the creation of units that require an offset, such as degrees Fahrenheit. This allows model authors to work in whatever set of units they feel most comfortable, while still ensuring that their models can be integrated with those of other authors using other units.
New units are defined using the <units> element, which may be placed inside both <model> and <component> elements. When a <units> element is placed inside the <model> element, the units definition may be referenced from within any component in the model. When a <units> element is placed inside a <component> element, the units definition may only be referenced from within that component.
Each units element must define a name attribute, which is used to reference the units definition elsewhere. The value of the name attribute must be unique across all <units> elements in the <model> or <component> element in which it is defined. If the value of the name attribute of a <units> element defined inside a <component> element matches the value of the name attribute on a <units> element defined inside the parent <model> element, then it will redefine the units, and all references to these units within the component element refer to the new definition. Model authors must not redefine any of the standard units. Therefore, the value of the name attribute must not equal one of the names from the standard units dictionary in Table 2.
A <units> element may also define a base_units attribute, the associated behaviour of which is discussed in Section 5.2.4. A <units> element can contain a set of <unit> elements that reference units from the dictionary or some previously defined units.
A <unit> element has no content but may have up to five attributes. The units attribute is the only one that is required. It is used to set the base quantity for the current <unit> element, and its value must correspond to a keyword from the standard CellML units dictionary or to the value of the name attribute of a <units> element in the current component or model.
The definition of new units in terms of subunits may require the use of some combination of the optional offset, prefix, exponent, and multiplier attributes.
A multiplier attribute can be used to pre-multiply the quantity to be converted by any real scale factor. For instance, a multiplier of 0.45359237 is used to define a pound in terms of a kilogram. The multiplier attribute has a default value of "1.0"
The offset attribute is used to represent the addition of a constant in the transformation between the current units and the base units. This should only be necessary for the definition of temperature scales. For instance, an offset attribute value of "32.0" is needed to define Fahrenheit in terms of Celsius. The offset attribute has a default value of "0.0".
The prefix attribute can be used to indicate a scale for the referenced units. It is included primarily for the convenience of modellers who want to define units that differ from another units definition only by an SI scale factor. Its value must be from the standard set of CellML prefix names given in Table 3 or be an integer, in which case the units are pre-multiplied by 10 to the power of this number. The default value of the prefix attribute is "0.0", (the referenced units are scaled by a factor of one).
| name | factor | symbol | name | factor | symbol |
|---|---|---|---|---|---|
| yotta | 1024 | Y | deci | 10-1 | d |
| zetta | 1021 | Z | centi | 10-2 | c |
| exa | 1018 | E | milli | 10-3 | m |
| peta | 1015 | P | micro | 10-6 | u |
| tera | 1012 | T | nano | 10-9 | n |
| giga | 109 | G | pico | 10-12 | p |
| mega | 106 | M | femto | 10-15 | f |
| kilo | 103 | k | atto | 10-18 | a |
| hecto | 102 | h | zepto | 10-21 | z |
| deka | 101 | da | yocto | 10-24 | y |
Table 3
The set of names that may be used in the prefix attribute on a <unit> element and the corresponding scale factors that will pre-multiply the unit. The standard abbreviation for each unit is provided for reference, but must not be used in the prefix attribute.
The scale factor described by the prefix attribute and the units referenced by the units attribute are raised to a power equal to the value of the exponent attribute. The value of the exponent attribute must be a floating point number, and is typically an integer. The exponent attribute has a default value of "1.0". Note that an exponent attribute value of "0.0" has the effect of removing the parent <unit> element from the current units definition.
A `simple units' definition occurs when units are defined as a linear function of some previously defined simple units or base units. This occurs when a <units> element contains only a single child <unit> element, that <unit> element has an exponent attribute value of "1.0", and the units definition referenced by the units attribute is one of the SI or user-defined base units or is itself a simple units definition. These are the only conditions under which a <unit> element may define an offset attribute. The formula that expresses how the old units (referenced by the value of the units attribute on the <unit> element) are transformed into the new units (defined by the value of the name attribute on the parent <units> element) is given below:
xnew[ Units ] = ( multiplier [ Units/units ] prefix ) xold [ units ] + offset [ Units ] (1)
Terms in square brackets represent the units associated with a term in the expression, xold is the value to be transformed from the old units, xnew is the resulting value in the new units, Units are the units being defined, and multiplier, prefix, units, and offset correspond to the values of the appropriate attributes on the <unit> element.
`Complex units' are the product of multiple base quantities, and are created by placing several <unit> elements inside a single <units> element, or by defining an exponent attribute with a value other than "1.0" on any <unit> element. The conversion between the new units and the product of the constituent units is given by the formula below:
xnew [ Units ] = xold [ u1e1 ... unen ] m1 [ Units1/n / u1e1 ] p1e1 ... mn [ Units1/n / unen ] pnen (2)
The mn, pn, un, and en terms refer to the values of the multiplier, prefix, units, and exponent attributes on the n-th <unit> element respectively. Note that this specification forbids offset attributes from being defined on any unit elements that occur inside a complex units definition.
It is very important to note that when a complex units definition references a simple units definition, any offset associated with the simple units definition is removed. This means that the conversions such as the one between degrees Fahrenheit per inch and degrees Celsius per centimetre involve only a scale factor.
5.2.4 New base units
A modeller might want to define and use units for which no simple conversion to SI units exist. A good example of this is pH, which is dimensionless, but uses a log scale. Ideally, pH should not simply be defined as dimensionless because software might then attempt to map variables defined with units of pH to any other dimensionless variables.
CellML addresses this by allowing the model author to indicate that a units definition is a new type of base unit, the definition of which can not be resolved into simpler subunits. This is done by defining a base_units attribute value of "yes" on the <units> element. This element must then be left empty. The base_units attribute is optional and has a default value of "no". If the base_units attribute is omitted or assigned a value of "no", units are expected to be defined in terms of other units as described in Section 5.2.3.
The indiscriminate use of the base_units attribute is strongly discouraged, because it has a significant impact on the re-usability of models and components. In particular, the base_units attribute should not be used to restrict users to creating models with an application-specific dictionary of units, as this prevents the efficient exchange of CellML models with other applications.
Software that is checking the consistency of the units in an equation (described in more detail in Section 5.2.5) can stop the recursive resolution of units definitions when the only remaining units are base SI units and user-defined base units.
5.2.5 Equation dimension checking
The association of units with every variable and bare number that appears in an equation in a CellML document provides CellML processing software the opportunity to perform equation dimension checking. Verifying that equations have consistent dimensions can potentially catch many basic mathematical errors. CellML Level One conformant software is free to ignore units in mathematics and assume that equations are consistent. CellML Level Two conformant software must check the consistency of dimensions in equations.
Section 5.5.5 specifies a possible implementation of equation dimensionality checking. This implementation splits an equation into a tree of equation parts, in which each parent part if obtained by the application of a single operator to its children. The units definition on each leaf node (i.e., part without children) is expanded into base units, as described in Section 5.5.4. The units definition for a node at a higher level of the tree is constructed by combining the units definition of its children. An equation has consistent dimensions if the fully expanded units definitions of the two nodes at the top level of the tree are equivalent, as defined in Section 5.5.3.
This specification does not require software to use the implementation discussed in Section 5.5.5, but does require that software that claims to perform dimension checking achieve the same results as if that implementation were used.
This specification does not attempt to completely prevent model authors from creating bad mathematics. Dimension consistency checking prevents modellers from adding variables with different dimensions but would not find errors in the following equations, which have different units but the same dimensions:
(x volts) = (y volts) + (z millivolts)
(x inches) = (y metres) + (z nautical_miles)
Although it would be technically possible to find and correct such errors, CellML processing software is not required to be able to do so.
5.2.6 Units and variable mapping
Associating units definitions with every variable declaration in a component allows variables from components that make use of different sets of units to be mapped together, as long as the variables have the same dimensions. Section 5.5.6 specifies a possible implementation of the conversion of a numeric value from one set of units to another. This specification does not require that software use this implementation but does require that software that claims to support units conversion during variable mapping achieve the same results as if this implementation were used.
This implementation generates an expression that relates each units definition to SI and user-defined base units. This expression is obtained by recursively expanding each units definition as described in Section 5.5.4, and then simplifying the result. The expression for the input units is then inverted to give an expression that relates the appropriate base units to the input units. This inverted expression is substituted into the expression for the target units, producing a single expression that relates the quantity to be converted from the input units to a corresponding quantity in the target units. The inversion and substitution process is described in Section 5.5.7.
5.3 Examples
5.3.1 Examples of user-defined units
Figure 8 shows several CellML units definitions, demonstrating how simple units can be expressed as linear functions of other simple units, and how complex units are obtained from the product of other units.
Figure 8
Some examples of the use of the <units> element demonstrating the definition of simple and complex units.
5.3.2 Examples of equation tree formation
The first step in the algorithm proposed in Section 5.5.5 for verifying that a given equation has consistent dimensions is to convert the equation into a tree of equation parts. A relational operator (typically the equals operator) combines the nodes at the top of the tree. For instance, the equation:
x = 3y(z + 2)
would have the tree shown in Figure 9.
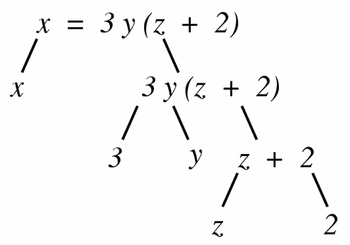
Figure 9
The tree form of the equation x = 3y(z + 2), in which each non-leaf node is obtained by the application of a single operator to its children.
5.4 Rules for CellML Documents
Units are a fundamental part of a CellML model definition. In this section, formal rules are specified for the system of units definition introduced in Section 5.2.
5.4.1 The <units> element
- Allowed use of the
<units>element-
Both the
<model>and<component>elements can contain any number of<units>elements. -
Each
<units>element must define anameattribute, and may define abase_unitsattribute. -
If a
<units>element defines abase_unitsattribute with a value of"yes", then that<units>element must contain only the following elements, which may appear in any order:- metadata framework elements, as described in Section 8.
-
If a
<units>element does not define abase_unitsattribute with a value of"yes", then that<units>element must contain only the following elements, which may appear in any order:.<unit>elements in the CellML namespace,- metadata framework elements, as described in Section 8.
-
Both the
- Allowed values of the
nameattribute-
The value of the
nameattribute must be a valid CellML identifier as discussed in Section 2.2.1. -
The value of the
nameattribute must not equal one of the names defined in the standard dictionary of units in Table 2. [ Model authors may not redefine the standard units. ] -
The value of the
nameattribute must be unique across all<units>elements at the same level in a CellML document. [ Two<units>elements in the same<model>element may not have the samenameattribute value, although a<units>element in a<component>element may share the same name as a<units>element in the parent<model>element. In this case, the units definition in the<component>element supercedes the model-wide definition when referenced inside that component. ]
-
The value of the
- Allowed values of the
base_unitsattribute-
If present, the value of the
base_unitsattribute must be"yes"or"no". -
If not present, the value of the
base_unitsattribute defaults to"no".
-
If present, the value of the
5.4.2 The <unit> element
- Allowed use of the
<unit>element-
A
<unit>element must contain only the following elements, which may appear in any order:- metadata framework elements, as described in Section 8.
-
Each
<unit>element must define aunitsattribute. It may also defineprefix,exponent,multiplier, andoffsetattributes.
-
A
- Allowed values of the
unitsattribute-
The value of the
unitsattribute must be taken from the standard dictionary of units listed in Table 2 or be the value of thenameattribute on a<units>element defined in the current<component>or<model>element. -
The value of the
unitsattribute must not reference a units definition that contains<unit>elements that in turn directly or indirectly reference the current units definition. [ This rule prevents circular units definitions. It must be possible to break down a complex units definition into the base SI units. ]
-
The value of the
- Allowed values of the
prefixattribute-
If present, the value of the
prefixattribute must be an integer or a name taken from one of the name columns of Table 3. [ The unit is scaled by 10 raised to the power of the specified integer or the factor corresponding to the specified name. Therefore,prefixattribute values of"centi"and"-2"are equivalent. ] -
If not present, the value of the
prefixattribute defaults to"0".
-
If present, the value of the
- Allowed values of the
exponentattribute-
If present, the value of the
exponentattribute must be a real number. -
If not present, the value of the
exponentattribute defaults to"1.0".
-
If present, the value of the
- Allowed values of the
multiplierattribute-
If present, the value of the
multiplierattribute must be a real number. -
If not present, the value of the
multiplierattribute defaults to"1.0".
-
If present, the value of the
- Allowed values of the
offsetattribute-
If present, the value of the
offsetattribute must be a real number. -
If not present, the value of the
offsetattribute defaults to"0.0".
-
If present, the value of the
- Proper use of the
offsetattribute-
A
<units>element containing a<unit>element that defines anoffsetattribute with a value other than"0.0"must not contain other<unit>elements. [ Theoffsetattribute can only be used in a simple units definition, as defined in Section 5.2.3. ] -
A
<unit>element that defines anoffsetattribute with a value other than"0.0"must not define anexponentattribute with a value other than"1.0". [ Theoffsetattribute can only be used in a simple units definition, as defined in Section 5.2.3. ]
-
A
5.5 Rules for Processor Behaviour
5.5.1 Resolving references to units definitions
The <units> element may be placed inside both <model> and <component> elements. When user-defined units are referenced by a variable or number declaration inside a component, the units definition is first looked for inside the current <component> element. If a matching units definition cannot be found, then the units definition is looked for in the parent <model> element.
5.5.2 Equivalence of units definitions
Two units references are considered identical if they satisfy one of the following criteria:
- They reference the same units definition from the standard dictionary.
- They reference the same units definition in the current
<component>element. - They reference the same units definition in the current
<model>element, where that units definition is not superceded by a units definition with the same name in the current<component>element.
5.5.3 Dimensional equivalence of units definitions
Two sets of units are considered equivalent if, when each is recursively resolved until left with nothing but products of SI and user-defined base units:
- the resolved form of each units definition consists of the same set of base units, and
- the exponent on each base unit is identical in each resolved units definition.
5.5.4 Expansion of units definitions
If software claims to perform dimension consistency checking of equations or conversion of units when mapping variables, it must obtain results that are equivalent to those produced using the algorithms described in Section 5.5.5 and Section 5.5.6, respectively. Both of these algorithms use the algorithm described in this section to expand units definitions into functions of the SI and user-defined base units.
This section derives a mathematical expression that relates units U to standard and user-defined base units. The specific steps in the derivation depend on whether the units definition for U is simple or complex, as defined in Section 5.2.3. Both derivations use recursive methods. At each step, any units that are not base units are replaced with expansions based on the appropriate definition.
The resolution of a simple units definition is straightforward, because the subunits on which the new units are based are also simple units. If units U are simple units, then the definition of U is given by:
xU[ U ] = ( m1 [ U/u1 ] p1 ) x1 [ u1 ] + o1 [ U ] (1)
where m1, o1, p1 and u1 are the values of the multiplier, offset, prefix, and units attributes on the <unit> element respectively, and u1 is another simple units definition given by:
x1[ u1 ] = ( m2 [ u1/u2 ] p2 ) x2 [ u2 ] + o2 [ u1 ] (2)
Equation 2 can be substituted into Equation 1 to give:
xU[ U ] = ( m1 [ U/u1 ] p1 ) ( ( m2 [ u1/u2 ] p2 ) x2 [ u2 ] + o2 [ u1 ] ) + o1 [ U ] (3)
Further levels of units definitions can be rearranged and resolved to simpler units as shown above until the resulting expression relates U to some base units. This final expression can be simplified to be in the following form.
xU[ U ] = mn [ U/un ] xn [ un ] + on [ U ] (4)
where un represents the final base units, and the constants mn and on are the result of the conversion of prefixes into scale factors according to the Table 3 and simplification.
For a complex units definition, the units U can be related to the subunits that are referenced in the definition using the following expression:
xU [ U ] = x1 [ u1e1 ... unen ] m1 [ U1/n / u1e1 ] p1e1 ... mn [ U1/n / unen ] pnen (5)
where mn, pn, un, and en refer to the values of the multiplier, prefix, units, and exponent attributes on the n-th <unit> element in the units definition respectively. Any units that appear in the expansion of the first units definition that are not base units should be expanded using the appropriate equation. The resulting expansion is then substituted directly into the parent expression in place of the relevant units reference. This expansion and substitution is performed recursively until the unit definitions referenced in the expression are base units.
It is very important to note that when a simple unit definitions is encountered in the expansion of a complex units definition, the fully expanded form of that simple units definition should be substituted into the parent expression without the constant offset term.
The final expansion can be simplified into the following form:
xU [ U ] = xn [ u1e1 ... unen ] mn [ U / u1e1 ... unen ] (6)
where mn is the multiplier resulting from the conversion of prefixes into scale factors according to the Table 3 and simplification, and un and en are the units name and corresponding exponent for the n-th base units.
Some examples of the expansion of units definitions will be made available soon.
5.5.5 Rules for equation dimension checking
If software chooses to verify that equations are self-consistent with respect to the dimensions of the units definitions referenced by all numbers and variables, it must obtain the same results as would be obtained by following these steps:
-
The equation is split into a tree of equation parts, in which each parent part is obtained by the application of a single operator to its children. A relational operator (typically the equals operator) combines the nodes at the top of the tree. An example of the tree formulation of an equation is given in Section 5.3.2. This specification will not attempt to further define this step.
- The units definitions for the terms at the leaves of the tree are expanded into functions of the SI and user-defined base units. The expansion of a units definition into base units is discussed in Section 5.5.4.
- Starting at the leaves of the tree, sets of child nodes can be recursively removed from the tree according to the operator applied to them. Operators such as addition or subtraction require that all of the child nodes have unit definitions with identical dimensions, as defined in Section 5.5.3. If this is true, the parent node assumes the same dimensions as its children. If it is not, the equation has inconsistent dimensions. There are no restrictions on the units definitions used by nodes combined using operators such as multiplication or division operators. The dimensions of the parent term assume the result of the appropriate operation on the dimensions of the child terms.
- The equation has self-consistent dimensions if the fully expanded units definitions of the two nodes at the top of the tree are equivalent, as defined in Section 5.5.3.
- It an inconsistency is detected at any point, then software is free to do whatever it likes. This specification recommends that it alert the user to a possible error, at the very least.
5.5.6 Units and variable mapping
If software claims to be able to perform units conversion when passing the value of a variable between components, it is required to produce results that are consistent with those that would be obtained using the algorithm described in this section.
If two variable declarations both reference identical units definitions as defined in Section 5.5.2, then there is a one-to-one mapping between the the variable's value in both components.
If two variable declarations reference different units definitions, some sort of units analysis and conversion is required to ensure that the model functions properly. If software chooses to perform this variable mapping, then it must be capable of converting any value of a variable x which is measured in U1 units to an equivalent value y measured in U2 units. This conversion must obtain the same results as would be obtained by following the procedure outlined below to derive a mathematical expression relating x to y:
-
Two mathematical expressions are generated, in which
U1andU2are functions of SI and user-defined base units. The method by which these expressions are generated is discussed in Section 5.5.4. -
U1andU2must have equivalent dimensions as defined in Section 5.5.3. -
The expression relating
U1to the base units is inverted, and combined with the expression forU2to give a single expression relatingxtoy. This inversion and substitution process is discussed in Section 5.5.7.
5.5.7 Generating an equation relating units definitions
The recursive resolution of units definitions according to the procedure defined in Section 5.5.4 leaves two equations that are of one of the following two forms:
znew[ U ] = m [ U/un ] zold [ un ] + o [ U ] (1)
znew [ U ] = zold [ u1e1 ... unen ] m [ U / u1e1 ... unen ] (2)
In most cases, both equations will be of the same form. If the expressions for U1 and U2 are of different forms, the two equations will only have equivalent dimensions if the expression of form shown in Equation 2 has a single base unit u1, and e1 has a value of one.
The expanded equations for U1 and U2 can be related by either un, if the expressions are of the form shown in Equation 1, or by the product u1e1 ... unen, if the equations are of the form shown in Equation 2. The equation for x can be inverted. This inversion will result in one of the forms shown below:
zold[ un ] = ( znew [ U ] - o [ U ] ) / m [ U/un ] (3)
zold [ u1e1 ... unen ] = znew [ U ] / mn [ U / u1e1 ... unen ] (4)
The inverted equation for x can be substituted into the equation for y to give a single equation defining y in terms of x. Examples of this procedure will be made available soon.
6 Grouping
6.1 Introduction
It is often useful to organise groups of components within a model into a hierarchical structure. This structure might reflect the logical organisation of components within the group or their physical configuration. CellML provides a single mechanism for the specification of both of these forms of hierarchy. This mechanism is based on a grouping scheme that allows model authors to create numerous hierarchical structures over a single network of components. The parent-child relationships in one hierarchical grouping need not necessarily be consistent with those specified in another grouping, a situation that could not be supported by nesting of component definitions.
It is anticipated that models will typically be defined as a network, with hierarchical relationships defined between groups of components at different places within the model. CellML processing software is free to treat these structures as discontinuous. Alternatively, it may combine structures that represent the same relationship into a single hierarchy by assuming that the root nodes of any hierarchical arrangements of components are all children of a single imaginary component. This imaginary component is not explicitly defined within the CellML document and has no properties.
The definition of a logical hierarchy of components in a network is known as "encapsulation". Encapsulation allows the modeller to hide a group of components from the rest of the model by using a single component as an interface to the hidden subnetwork. The parent component hides the details of one or more child components from the rest of the model. Encapsulation provides a powerful mechanism for simplifying the structure of the model by preventing connections between specified sets of components. Components in the main network may not connect to the child components in the subnetwork — all variables must be mapped through the parent interface component. Components in the subnetwork may only be connected to the interface component and to other components in the same subnetwork, which may include further levels of encapsulation. Therefore, a modeller wishing to re-use an encapsulated subnetwork may treat the subnetwork as a "black box", and deal exclusively with the interface presented by the encapsulating component.
The definition of physical hierarchies within a model is known as "containment" in CellML. A model author can specify that one or more child components are physically inside of a parent component without describing the geometric aspects of the relationship in detail. This information would typically be used by CellML processing software to provide simple renderings of a model.
Model authors are also free to extend the grouping scheme with user-defined types of relationships between components. However, CellML processing software is only expected to recognise encapsulation and containment relationships.
Groups do not add any additional mathematical information to the model. Model authors may not define their own grouping relationships that are intended to imply mathematical information.
6.2 Basic Structure
6.2.1 Definition of groups
Logical and physical hierarchies are both declared using the <group> element. This element must be a child of a <model> element. Each <group> element contains one or more <relationship_ref> elements, each of which defines a relationship attribute, the value of which references the type of relationship represented by the group. CellML processing software is expected to recognise two types of relationship: encapsulation and containment, which are indicated by relationship attribute values of "is_encapsulated_by" and "is_contained_in", respectively.
The <group> element also contains two or more <component_ref> elements, each of which defines two attributes. The component attribute references a component within the current model. The role attribute indicates whether the component is the dominant component in the hierarchy. A component referenced by a <component_ref> element with a role attribute value of "major" is the dominant component. This component is the encapsulating component in a logical encapsulation hierarchy or the containing component in a geometric containment hierarchy. A <group> element contains one or more <component_ref> elements with a role attribute value of "minor". The components referenced by these elements are the encapsulated components in a logical encapsulation hierarchy or the contained components in a geometric containment hierarchy. A <group> element that defines a logical encapsulation or geometric containment relationship must reference exactly one major component and at least one minor component. A group element that defines a user-defined type of relationship may have any number of minor and major components.
A single <group> element may be used to define multiple relationships between components. For instance, encapsulation and geometric relationships may be defined within the same <group> element and thus share the same hierarchy. This is done by including more than one <relationship_ref> element in the <group> element. Each <relationship_ref> element must define a relationship attribute, which may be in the CellML namespace or in an extension namespace. The value of the relationship attribute names the type of relationship referenced by the <relationship_ref> element. If the relationship attribute is in the CellML namespace, its value must be either "is_encapsulated_by" or "is_contained_in". A <relationship_ref> element may also define a name attribute. The value of the name attribute on <relationship_ref> elements can be used to combine several <group> elements into a single hierarchical structure (see Section 6.2.4 for more information on this).
Geometric containment relationship information is formally independent of logical encapsulation information, but CellML processing software is free to check for inconsistencies between the two relationships — it would generally not be useful for an encapsulating component to be physically inside one of its encapsulated child components.
All children of a given major component in a single hierarchy must appear within a single <group> element. This simplifies the construction and validation of hierarchies from <group> elements. For instance, the requirement that a component may only have a single parent in any given hierarchy would be difficult to enforce if minor components could be scattered across several <group> elements.
6.2.2 The encapsulation relationship
Encapsulation allows the modeller to split a model into layers of complexity. A single component can be used to encapsulate a complex partial model, and thereby provide a unified interface for all information passing between that subnetwork and the rest of the model.
A model may only define a single encapsulation hierarchy, which may be continuous or discontinuous. Each component in the hierarchy may have at most one parent component. If the hierarchy is continuous, the parent component will always be another component defined within the current model. If the hierarchy is discontinuous, it may be convenient to assume that any unencapsulated components are children of a single imaginary component. This imaginary component makes it easier to check that the hierarchy has no circular relationships between components.
The components in a model can be divided into four sets with respect to any given component (the current component). The set of all components immediately encapsulated by the current component is the encapsulated set. The parent component is the component that encapsulates the current component. Other components encapsulated by the same parent make up the sibling set. All other components, which are not available to make connections with the current component, make up the hidden set. If the current component is not encapsulated, then it has no parent and the sibling set consists of all other unencapsulated components in the model.
These sets are best demonstrated by example. Given the network shown in Figure 10, Table 4 lists the parent components and the components in the encapsulated, sibling, and hidden sets for a selected set of components picked as the current component.
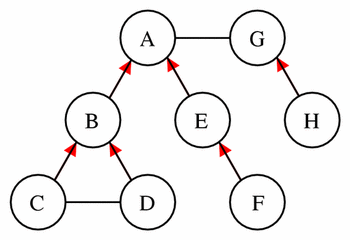
Figure 10 This simple model provides the basis for the demonstration of the concepts of encapsulated sets, parents, sibling sets, and hidden sets, as described in the text. The model consists of eight components each represented by a circle. The lines between the components represent connections, and a red arrowhead on one of these lines indicates that the component at the tail of the arrow is encapsulated by the component at the head of the arrow.
| Current Component | Encapsulated Set | Parent | Sibling Set | Hidden Set |
|---|---|---|---|---|
| A | B, E | imaginary | G | C, D, F, H |
| B | C, D | A | E | F, G, H |
| C | none | B | D | A, E, F, G, H |
| E | F | A | B | C, D, G, H |
| G | H | imaginary | A | B, C, D, E, F |
Table 4 This table lists the parent components, and the components in the encapsulated, sibling, and hidden sets for a selected few components from the example model in Figure 10. Components A and G do not have a real parent component, but may have an imaginary parent component that enables the formation of a single encapsulation hierarchy.
Every variable must define its availability for use in other components. This is done with the public_interface and private_interface attributes on the <variable> element. The interface exposed to the parent component and components in the sibling set is defined by the public_interface attribute. The private_interface attribute defines the interface exposed to components in the encapsulated set. Each interface has three possible values: "in", "out", and "none", where "none" indicates the absence of an interface. The separation of interfaces allows the modeller to incrementally add complexity to a encapsulated network without changing the interface presented to the rest of the network by the encapsulating component.
The mappings that are allowed between variables declared in each component are controlled by the public and private interfaces of each variable and the prohibition on connecting an encapsulated component to components other than its parent component, members of its sibling set, and any components it in turn encapsulates. Variables with a public_interface attribute value of "in" must be mapped to a single variable in the sibling set with a public_interface attribute value of "out" or to a single variable in the parent of the current component with a private_interface attribute value of "out". Similarly, variables with a public_interface value of "out" may be mapped to variables in components in the sibling set with a public_interface attribute value of "in" or to variables in the parent component with a private_interface value of "in". Note that defining a public_interface attribute value of "out" on a variable makes it legal to map the variable to other variables, but does not require that such a mapping occur. If a variable has a public_interface attribute value of "none", it cannot be mapped to variables in the parent component or to variables in components in the sibling set.
Variables with a private_interface attribute value of "in" must be mapped to a single variable from a single component in the encapsulated set with a public_interface attribute value of "out". Variables with a private_interface attribute value of "out" may be mapped to any variables from components in the encapsulated set with a public_interface attribute value of "in". If a variable has a private_interface attribute value of "none", it is neither input from or exposed to the components in the encapsulated set.
If both the public_interface and private_interface attributes of a variable have a value of "none", the variable can only be used in the current component and is invisible to all other components in the model. In order to determine which variables may be modified in the current component, we must determine if either the public_interface attribute or the private_interface attribute has a value of "in". If so, the variable is declared elsewhere and its value may not be mathematically modified in the current component. If not, the variable belongs to the current component.
The two interface attributes of a variable are completely independent with one exception: it is invalid for a variable to have both public_interface and private_interface attributes with value "in". An interface with value "in" reflects an unmet need in the current component that must be satisfied — this need can be met in either the public or private interface, but not both.
6.2.3 The containment relationship
The is_contained_in relationship allows the modeller to specify that a particular component is physically inside another. This might be used by software for the rendering of a model. Containment relationships can be specified either in combination with or independent of encapsulation relationships. Containment relationships do not restrict any aspect of model definition or behaviour.
6.2.4 Named containment hierarchies
CellML allows the definition of multiple containment hierarchies over the same network model. This functionality allows the modeller to define several different ways of organising the same model, each of which might highlight a different aspect of the model's physical structure. This functionality has been included in CellML for extended compatibility with AnatML, an XML-based language for describing anatomical structures.
A containment hierarchy is created when several <group> elements contain <relationship_ref> elements with a relationship attribute value of "is_contained_in" and the same name attribute value. Any <group> elements that contain <relationship_ref> elements with a relationship attribute value of "is_contained_in" and that do not define a name attribute are also considered to form a single grouping hierarchy.
As was the case for encapsulation grouping, a containment hierarchy may be continuous or discontinuous. Each component in the hierarchy may have at most one parent component. If the hierarchy is continuous, the parent component will always be another component defined within the current model. If the hierarchy is discontinuous, it may be convenient to assume that any components not already contained within other components are children of a single imaginary component. This imaginary component makes it easier to ensure that the hierarchy has no circular relationships between components.
6.2.5 User-defined relationship types
Modellers are free to use the grouping syntax of CellML to organise model components in ways not described in the CellML specification. To do this, the model author defines a new relationship type, the name of which is used as the value of the relationship attribute on the <relationship_ref> element. The relationship attribute must be placed in an extension namespace, because future versions of the CellML specification may define additional relationship types, the names of which could otherwise conflict with user-defined relationship types. If a modeller uses a non-standard value for the relationship attribute, the value used should indicate the relationship between minor and major components. A <group> element that defines a user-defined type of group is free to contain only minor components. For example, a modeller may define a grouping class called "is_next_to", used to tell a processor that one minor component is physically adjacent to another.
Modellers are free to use the name attribute on the <relationship_ref> element to specify multiple hierarchies for user-defined relationship types, as is possible for the containment relationship.
This specification does not provide a mechanism by which modellers may specify the meaning of a user-defined type of relationship. This definition must be provided by the processing software declaring the new relationship type.
6.3 Examples
Figure 11 demonstrates the use of the <group> element to define an encapsulation relationship. This example is taken from the two reaction pathway with encapsulation example from the examples section of the CellML website. It shows how a component representing an overall reaction (total_reaction) can encapsulate components representing intermediate reactions (first_reaction and second_reaction) and their by-products (C and D).
Figure 11
Example demonstrating the use of the <group> element to define a logical encapsulation relationship. See text for more details.
Figure 12 demonstrates the use of the <group> element to define encapsulation and containment relationships, the construction of two named geometric hierarchies, and the specification of a custom relationship type (is_next_to) in an extension namespace. Most CellML models will probably only define a single geometric hierarchy. In this case, it is not necessary to name the hierarchy, since all unnamed groups are assumed to belong to the same geometric hierarchy.
Figure 12
Examples demonstrating the use of the <group> element. See text for more details.
The first <group> element states that the cell_membrane component is physically inside the cell component, and that this geometric relationship is part of a geometric hierarchy called membrane. The second <group> element states that the sodium_channel and calcium_channel components are both physically inside and logically encapsulated by the cell_membrane component. This completes the membrane geometric hierarchy. The encapsulation relationship prevents the sodium and calcium channel components from being connected to any components other than the cell_membrane component, each other, and any components they in turn encapsulate.
The third <group> element states that the two components representing parts of the sarcoplasmic reticulum are physically inside the cell, and that this relationship is part of a geometric hierarchy called intracellular. Finally, the fourth <group> element introduces the user-defined relationship is_next_to, and states that the the two sarcoplasmic reticulum components share this relationship. This relationship type is declared by putting the relationship attribute in an extension namespace, and assigning it a value of "is_next_to". Note that this relationship has no major or dominant component, and that CellML processing software is free to ignore the information provided by this group.
6.4 Rules for CellML Documents
6.4.1 The <group> element
- Allowed use of the
<group>element-
A
<model>element may contain any number of<group>elements. -
A
<group>element must contain only the following elements, which may appear in any order:-
<relationship_ref>and<component_ref>elements in the CellML namespace, - metadata framework elements, as described in Section 8.
<group>element in the order stated above. ] -
-
A
<group>element must contain at least one<relationship_ref>element. -
A
<group>element must contain at least one<component_ref>element.
-
A
6.4.2 The <relationship_ref> element
- Allowed use of the
<relationship_ref>element-
A
<relationship_ref>element must contain only the following elements, which may appear in any order:- metadata framework elements, as described in Section 8.
-
Each
<relationship_ref>element must define arelationshipattribute in either the CellML namespace or an extension namespace. It may also define anameattribute. [ Arelationshipattribute declaring a user-defined relationship type is placed in an extension namespace. This prevent conflicts with future versions of the CellML specification, which may define additional types of relationships in the CellML namespace. ]
-
A
- Allowed values of the
relationshipattribute-
The value of a
relationshipattribute in the CellML namespace must be"is_contained_in"or"is_encapsulated_by".
-
The value of a
- Allowed values of the
nameattribute-
The value of the
nameattribute must be a valid CellML identifier as discussed in Section 2.2.1. [ Note that unlike most othernameattributes, the value of thenameattribute on a<relationship_ref>element is not expected to be unique across the current model. Instead,<group>elements that include<relationship_ref>elements that share the samenameattribute value form are parts of a single hierarchy. ]
-
The value of the
- Proper use of the
nameattribute-
A
nameattribute may not be defined on a<relationship_ref>element with arelationshipattribute value of"is_encapsulated_by". [ A model may define only a single, unnamed encapsulation hierarchy. ]
-
A
6.4.3 The <component_ref> element in <group> elements
- Allowed use of the
<component_ref>element within a<group>element-
A
<component_ref>element must contain only the following elements, which may appear in any order:- metadata framework elements, as described in Section 8.
-
A
<component_ref>element within a<group>element must define acomponentattribute and aroleattribute.
-
A
- Proper use of the
<component_ref>element in<group>elements-
Two
<group>elements that contain<relationship_ref>elements with identicalrelationshipattribute values and undefinednameattributes may not reference the same major component. [ A single level of a hierarchy must only be defined with a single<group>element. It would be much more difficult to assemble a hierarchy from a CellML document if a single level of the hierarchy could be shared among multiple<group>elements. ] -
Two
<group>elements that contain<relationship_ref>elements with identicalrelationshipandnameattribute values may not reference the same major component. [ This rule extends the previous rule to include named hierarchies. ] -
A component must not be referenced as a minor component more than once in a single grouping hierarchy. All
<component_ref>elements with a commoncomponentattribute value and aroleattribute value of minor must be in different hierarchies. [ A grouping hierarchy must not be circular. ]
-
Two
- Allowed values of the
componentattribute-
The value of the
componentattribute must equal the value of thenameattribute of a<component>element contained within the current<model>element. -
The value of the
componentattribute on a<component_ref>element must be unique across all<component_ref>elements within the parent<group>element. [ A component may only appear once within a group. ]
-
The value of the
- Allowed values of the
roleattribute-
The value of the
roleattribute on a<component_ref>element in a<group>element must be either"major"or"minor".
-
The value of the
- Proper use of the
roleattribute-
A
<group>element that contains a<relationship_ref>element with arelationshipattribute of"is_encapsulated_by"or"is_contained_in"must contain exactly one<component_ref>element with aroleattribute value of"major"and at least one<component_ref>element with aroleattribute value of"minor". [ Groups defining an encapsulation or containment relationship must have exactly one dominant component and at least one minor component. ]
-
A
6.5 Rules for Processor Behaviour
6.5.1 Allowing multiple grouping hierarchies in a single model
A given model may define multiple geometric containment hierarchies, but may only define one logical encapsulation hierarchy.
A grouping hierarchy is built up from multiple <group> elements based on the value of the name attribute of the <relationship_ref> elements. All <group> elements that contain <relationship_ref> elements that share the same relationship and name attribute values are considered to form a single grouping hierarchy. All <group> elements that contain <relationship_ref> elements that share the same relationship attribute value and do not define name attributes are also considered to form a single grouping hierarchy.
If, after the groups that make up a single hierarchy are assembled, the resulting hierarchy is discontinuous, it may be convenient to assume that any components that are not already children of other components are children of a single imaginary component. The imaginary component has no properties in the model. Its sole purpose is to make it easier to check that the hierarchy has no circular relationships between components.
6.5.2 Groups must not imply mathematical information
Modellers are explicitly forbidden from using CellML groups to add mathematical information to the model. Modellers may not define their own types of relationships that imply mathematics.
6.5.3 Groups should not imply metadata information
Modellers should not use CellML groups to associate properties or classification information with sets of components. The metadata functionality is the proper method for making such associations. This increases the chance of that information being used by a range of CellML processing software.
7 Reactions
7.1 Introduction
CellML is intended to be used to represent many different types of models. Therefore, its basic structure is rather general, and models are primarily specified by explicitly defining mathematics using MathML. It will always be possible to specify a model purely in terms of mathematics, without using any of the elements defined in this section of the specification. However, in some types of models, information is lost in reducing the model to pure mathematics. For instance, in biochemical pathway models it will not always be straightforward, or even possible, to unambiguously determine from the mathematical rate laws which variables represent inhibitors or activators in the reactions. Therefore, some additional elements were needed in CellML to fully capture the information in biochemical pathway models.
7.1.1 Pathway model representations supported by CellML
Three fundamental representations of reaction/pathway models must be supported by CellML:
- Mathematical Equations: these are any valid mathematical equations that describe the model. For example, they may be ordinary differential equations that define kinetic reaction rate laws and the rate of change of the concentration of species participating in the modelled reactions.
-
Chemical Expressions: these are the stoichiometric expressions (such as
A + B <-> 2C + D) used by chemists to represent reactions. - Pathway Diagrams: these are the stylised drawings commonly used by biochemists and cell biologists to represent interactions among participants in reactions. Some examples of pathway diagrams are shown in Section 7.3.
It is important that CellML be able to store the information needed to unambiguously reproduce any of these representations of a model. It is also important to minimise duplication of information within the model definition, because duplication can lead to inconsistencies. Therefore, we must integrate the information needed to support the three types of model representation.
The integration process has resulted in the introduction of a CellML syntax that implies a mathematical relationship between variables in the current component. In this section of the specification, explicit mathematics refers to equations defined using MathML, and implicit mathematics refers to equations implied from the CellML syntax.
7.1.2 Qualitative vs. quantitative pathway models
CellML supports both quantitative and qualitative pathway models. Many types of models are commonly referred to as "qualitative". Some of these are mathematically specified, while others are not. For the purposes of this specification, qualitative pathway models consist solely of information about how the different chemical species in the pathway relate, and contain no mathematics. However, the stoichiometry of the reactions may be known. In other words, there is no mathematical representation of the model, but there may still be a pathway diagram and chemical expressions that represent the model. Because there is no mathematics in a qualitative model, CellML processing software is not required to be able to run a simulation using a qualitative model. However, some software may support simple simulations using such models.
Any model in which the change of concentration of a chemical species participating in a reaction is implicitly or explicitly defined is quantitative. All others are qualitative.
7.2 Basic Structure
The <reaction> element is used to store information associated with a single reaction. It may only appear inside of a <component> element. Examples of the <reaction> element are shown in Section 7.3. It is possible for a single <component> element to contain more than one <reaction> element. However, this practice makes it more difficult to re-use the individual reactions, and is therefore not recommended. The <reaction> element may define a reversible attribute, the value of which indicates whether or not the reaction is reversible. The default value of the reversible attribute is "yes".
The reaction element contains multiple <variable_ref> components, each of which references one of the variables that participates in the reaction. The recommended practice is to create a <variable_ref> element for each variable representing the concentration of a chemical species that participates in a reaction, as well as one for the variable representing the rate of the reaction. The required variable attribute is the only attribute on the <variable_ref> element. Its value is the name of the referenced variable. This variable must be declared in the current <component> element.
Each <variable_ref> element contains one or more empty <role> elements. There are four possible attributes on the <role> element. The required role attribute specifies the way in which the variable participates in the reaction. There are currently seven values allowed for this attribute: "reactant", "product", "catalyst", "activator", "inhibitor", "modifier", and "rate". These are defined in Section 7.4. The optional direction attribute should only be used on <role> elements in reversible reactions. It may have values of "forward", "reverse", or "both" and indicates the direction of the reaction for which the role is relevant. It has a default value of "forward". The optional delta_variable attribute indicates which variable is used to store the change in concentration of the species represented by the variable referenced by the current <variable_ref> element. The optional stoichiometry attribute stores the stoichiometry of the current variable relative to the other reaction participants. Section 7.4 contains detailed rules for the use of these attributes.
The <role> elements may also contain <math> elements, which define equations using MathML. Although it is not required, it is recommended practice to store all of the equations that relate to a reaction inside the appropriate <role> elements in the <reaction> element. This makes the <reaction> element more re-usable. In addition, defining mathematics inside a <role> element has the effect of associating the equations with the variable referenced by the containing <variable_ref> element, in the role defined by the <role> element. This enables CellML processing software to present the equations in a more meaningful context. For instance, it might group all of the relationships between the rate variable and the delta variables for all of the reactants and products, or it might display these equations in a different color. (Note that CellML processing software is not required to provide such additional functionality.)
There are three uses for equations inside <role> elements:
-
If the
roleattribute value is"rate", any enclosed equations calculate the kinetic rate law (i.e., calculate the value of the referenced variable) and the value of intermediate variables used in the rate law equation. -
If the
roleattribute value is"reactant"or"product", the equations calculate the relationship between the general reaction rate and the rate of change of the species represented by the referenced variable (i.e., calculate the value of the variable named in thedelta_variableattribute), and calculate any intermediate variables used in this relationship. -
In all other cases, the equations relate an intermediate variable used in the rate calculation to the variable referenced by the containing
<variable_ref>element. For instance, it would be appropriate to calculate an effective concentration of a catalyst inside the<role>element contained by the<variable_ref>element that references the variable representing the actual concentration of the catalyst.
Note that CellML processing applications are not required to be able to deduce the stoichiometry of a reaction from explicit mathematics. Therefore, it is strongly recommended that the stoichiometry and delta_variable attributes be used instead of explicit mathematics if the concentration change is simply the reaction rate multiplied by the stoichiometry. (The rules for deriving this mathematical relationship from the stoichiometry attribute are defined in Section 7.5.3.)
7.3 Examples
This section contains two examples demonstrating the recommended use of the <reaction> and <role> elements to define two basic reactions. The mathematics defining the reaction rate have been omitted in these examples. See the reaction model examples section of the CellML website for further examples.
Figure 13 shows a pathway diagram representation of the following reversible reaction:
A + B <-> 2C + D
Figure 14 demonstrates the use of CellML to define this reaction. There are five <variable_ref> elements in the <reaction> element: one for each variable representing the concentration of a chemical species participating in the reaction, and one for the variable representing the general reaction rate. Note that the stoichiometry attribute has a value of "2" for the variable representing the chemical species C, since this species appears with a stoichiometry of 2 in the chemical expression. The reversible attribute on the <reaction> element and the direction attributes on the <variable_ref> elements have their default values ("yes" and "forward", respectively) and therefore could have been omitted. They are included for clarity.
Figure 13
A typical pathway diagram representation of the simple reversible reaction A + B <-> 2C + D.
Figure 14
The CellML definition of the simple reversible reaction A + B <-> 2C + D. See text for more details.
Figure 15 shows the pathway diagram for the following irreversible, catalyzed reaction, which exhibits product-inhibition:
A + B -> D (catalyzed by C, inhibited by D)
The CellML definition of this reaction is shown in Figure 16.
Figure 15
A typical pathway diagram representation of the irreversible reaction A + B -> D (catalyzed by C, inhibited by D).
Figure 16
The CellML definition of the irreversible reaction A + B -> D (catalyzed by C, inhibited by D). See text for more details.
The <variable_ref> element that references the variable representing the concentration of species D now contains two <role> elements, one with information about D as a product and the other with information about D as an inhibitor. In this example, D has the same stoichiometry in both roles, but this would not necessarily need to be the case.
7.4 Rules for CellML Documents
7.4.1 The <reaction> element
- Allowed use of the
<reaction>element-
A
<component>element may contain any number of<reaction>elements. [ The use of multiple<reaction>elements within a single<component>element is discouraged, but is not illegal. ] -
A
<reaction>element must contain only the following elements, which may appear in any order:-
<variable_ref>elements in the CellML namespace, - metadata framework elements, as described in Section 8.
<variable_ref>element for each variable representing a chemical species that participates in the reaction, and one<variable_ref>element for the variable representing the rate of the reaction. ] -
-
The
<reaction>element may define areversibleattribute.
-
A
- Allowed values of the
reversibleattribute-
If present, the
reversibleattribute must have a value of"yes"or"no". -
If not present, its value defaults to
"yes". [ It is recommended to always explicitly define the value of this attribute. ]
-
If present, the
- Proper use of the
<reaction>element in encapsulating components[ It is often convenient to include a
<reaction>element in a component that is encapsulating several intermediate reactions (see Section 6 for more information about encapsulation). The encapsulating component represents an overall, or total, reaction, which can be represented by a<reaction>element. This total reaction is effectively qualitative, because any mathematics representing the progression of the total reaction is defined in the components representing the intermediate reactions. ]-
A
<reaction>element in an encapsulating component may not containdelta_variableattributes on the<role>elements or explicit mathematics defining the overall reaction rate or the changes in concentration of the species that participate in the total reaction. [ A valid CellML model must not define an inconsistent set of equations. Therefore, one should not introduce explicit or implicit mathematics in an encapsulating component that duplicates or contradicts mathematics (either explicit or implicit) defined in the encapsulated components. ]
-
A
7.4.2 The <variable_ref> element within a <reaction> element
- Allowed use of the
<variable_ref>element within a<reaction>element-
A
<variable_ref>element in a<reaction>element must contain only the following elements, which may appear in any order:<role>elements in the CellML namespace,- metadata framework elements, as described in Section 8.
-
Each
<variable_ref>element within a<reaction>element must contain at least one<role>element. [ The recommended best practice is to define one<role>element for each role assumed by the chemical species represented by the referenced variable. ] -
Each
<variable_ref>element within a<reaction>element must define avariableattribute.
-
A
- Allowed values of the
variableattribute-
The value of the
variableattribute on a<variable_ref>element within a<reaction>element must equal the value of thenameattribute on a<variable>element defined inside the current<component>element. -
The value of the
variableattribute must be unique across all<variable_ref>elements contained within the parent<reaction>element. [ A variable may only be referenced once in a single reaction. ]
-
The value of the
7.4.3 The <role> element
- Allowed use of the
<role>element-
A
<role>element must contain only the following elements, which may appear in any order:-
<math>elements in the MathML namespace, - metadata framework elements, as described in Section 8.
<role>elements are provided below, and rules for the<math>element and its children are given in Section 4. ] -
-
Each
<role>element must define a validroleattribute value. It may also definedirection,delta_variable, andstoichiometryattributes, subject to the constraints specified in the subsequent sections.
-
A
- Allowed values of the
roleattribute-
The
roleattribute must take one of the following seven values:-
"reactant": the species represented by the referenced variable is one of the species consumed or transformed by the reaction (in the forward direction). Reactants are also often called substrates. -
"product": the species represented by the referenced variable is one of the species produced by the reaction (in the forward direction). -
"catalyst": the species represented by the referenced variable catalyzes the reaction. In biochemical pathways, such a species will almost always be an enzyme and will almost always occur with astoichiometryattribute value of"1". -
"activator": the species represented by the referenced variable enhances the reaction. Activators can occur with any stoichiometry. An activator will usually be a small molecule that increases the activity of an enzyme catalyzing the reaction. However, the detailed reaction representing this activation of the enzyme may not be included in the model. Instead, the activator may be represented as directly affecting the kinetics of the catalyzed reaction. -
"inhibitor": the species represented by the referenced variable inhibits the reaction. Inhibitors can occur with any stoichiometry. An inhibitor will usually be a species that inhibits the activity of an enzyme catalyzing the reaction. However, the detailed reaction representing this inhibition of the enzyme may not be included in the model. Instead, the inhibitor may be represented as directly affecting the kinetics of the catalyzed reaction. -
"modifier": the species represented by the referenced variable modifies the reaction in some unspecified way. -
"rate": the referenced variable represents the rate of the reaction.
-
-
The
- Proper use of the
roleattribute-
Only one
<variable_ref>element in a given<reaction>element can contain a<role>element with aroleattribute with a value of"rate". [ There may only be one rate variable per reaction. ] -
A
<variable_ref>element that contains a<role>element with aroleattribute value of"rate"must not contain other<role>elements. [ The variable assigned the"rate"role may not be assigned any other roles. ] -
A
<role>element with aroleattribute of"rate"may not also definedirection,delta_variable, orstoichiometryattributes. [ The reaction rate should always be defined in the forward direction. To do otherwise will cause the implicit mathematics defined by thedelta_variableandstoichiometryattributes of the reactant and product roles to be erroneous. Thedelta_variableandstoichiometryattributes have no meaning for a rate variable. ] -
If a
<role>element has aroleattribute value of"reactant", there must be no other<role>element within the same parent<variable_ref>element with aroleattribute value of"product". [ A species may not be explicitly defined to be both a product and a reactant, although this is implied by a reversible reaction. ]
-
Only one
- Allowed values of the
directionattribute-
If present, the
directionattribute must take one of the following three values:-
"forward": the value of theroleattribute is the role of the referenced variable in the reaction when running in the "favoured" direction. The favoured direction is the one in which the the reactants are being consumed (i.e., the time-derivatives of their concentrations are negative), as defined by the kinetic rate law. -
"reverse": the value of theroleattribute is the role of the referenced variable in the reaction when running opposite to the "favoured" direction. In this direction, the reactants (as defined by the kinetic rate law) are being produced. -
"both": the value of theroleattribute is the role of the referenced variable in both directions of the reaction.
-
-
If not present, the value of the
directionattribute defaults to"forward".
-
If present, the
- Proper use of the
directionattribute-
A
directionattribute must only be defined on<role>elements contained in a<reaction>element on which thereversibleattribute has a value of"yes". [ Only reversible reactions may occur in two directions. ] -
The
directionattribute on a<role>element for which theroleattribute has a value of"reactant"or"product"must only have a value of"forward". [ This prevents the definition of inconsistent chemistry that could occur if a species could be explicitly defined as both a reactant and a product. ] -
The
directionattribute must only assume the value of"both"on<role>elements with aroleattribute value of"catalyst","activator","inhibitor", or"modifier". [ It is not chemically sensible to say that a species is a"reactant"in both directions. Nor does it make sense to declare that a species is a"product"in both directions. ] -
Each
<role>element contained in a given<variable_ref>element must have a unique combination of values for theroleanddirectionattributes. [ Defining two<role>elements with the sameroleanddirectionattribute values would allow the definition of inconsistent stoichiometries or multiple delta variables for a single variable. Both of these situations would create invalid CellML. ]
-
A
- Allowed values of the
stoichiometryattribute-
If present, the value of the
stoichiometryattribute must be a real number. [ In most cases, the value will be an integer. However, a valid CellML model may use fractional stoichiometries. ] -
The absence of a
stoichiometryattribute formally implies nothing. [ The absence of a stoichiometry value specifically does not imply a stoichiometry of"1". Instead, it would usually mean that the stoichiometry of the reaction is unknown. ]
-
If present, the value of the
- Allowed values of the
delta_variableattribute-
If present, the value of the
delta_variableattribute must equal thenameattribute on a<variable>element defined inside the current<component>element. -
The absence of the
delta_variableattribute implies nothing. -
The value of the
delta_variableattribute must be unique across all<role>elements contained within the parent<component>element. [ One variable cannot represent the rate of change in concentration of more than one species. The value of thedelta_variableattribute must be unique across the entire<component>element because it is legal (but not recommended) to include more than one<reaction>element in a single component. ]
-
If present, the value of the
- Proper use of the
delta_variableattribute-
The
delta_variableattribute may only appear on<role>elements in which theroleattribute equals"reactant"or"product". [ It is only in these roles that a chemical species may undergo a change in concentration. ] -
A
<role>element on which adelta_variableattribute is declared must also either declare astoichiometryattribute or include at least one<math>element in the MathML namespace. [ The combination of thedelta_variableattribute and thestoichiometryattribute implies a mathematical relationship between the variable referenced in thedelta_variableattribute and the variable assigned the role of"rate", as defined in Section 7.5.3. If thestoichiometryattribute is absent, the relationship between the variable named in thedelta_variableattribute and the variable assigned the role of"rate"must be defined using MathML. ] -
A
<role>element on which thestoichiometryanddelta_variableattributes are both defined must not also include<math>elements in the MathML namespace. [ The equations in a<math>element inside a<role>element for which theroleattribute is"reactant"or"product"must relate the variable named in thedelta_variableattribute to the variable assigned the role of"rate". Such equations would contradict the relationship implied by thedelta_variableandstoichiometryattributes, as defined in Section 7.5.3. ] -
If the
delta_variableandstoichiometryattributes are both declared on any single reaction participant, a<variable_ref>element must be provided for the variable that represents the reaction rate. This<variable_ref>must contain exactly one<role>element, with aroleattribute equal to"rate". [ Note that the reverse is not true: a variable may be assigned a role of"rate"even if the"reactant"and"product"variables do not definedelta_variableattributes. In this case, the modeller may choose to provide explicit mathematics relating the"rate"variable to the change in concentration of the various chemical species. ]
-
The
- Proper use of a
<math>element inside a<role>element-
A
<math>element in the MathML namespace inside a<role>element must define equations that are relevant to the variable referenced by the containing<variable_ref>element, acting in the role defined by theroleattribute on the<role>element. [ The meaning of "relevant" in this context is discussed in Section 7.5.4. ]
-
A
7.5 Rules for Processor Behaviour
7.5.1 Implications of the reversible attribute on the <reaction> element
If the reversible attribute has a value of "yes", it is assumed that all reactants in the forward direction are products in the reverse direction and vice versa. Similarly, all products in the forward direction are assumed to be reactants in the reverse direction and vice versa.
7.5.2 Chemical information implied by the stoichiometry attribute
The value of the stoichiometry attribute on a <role> element is defined to be the stoichiometry of the chemical species whose concentration is represented by the variable referenced by the containing <variable_ref> element. This stoichiometry can be used to produce the chemical expression representation of the model.
7.5.3 Math implied by the delta_variable and stoichiometry attributes
The use of the delta_variable and stoichiometry attributes on a <role> element implies the following mathematical relationship between the declared delta variable and the rate variable:
-
For reactants:
delta_variable = (stoichiometry)(rate) -
For products:
delta_variable = -(stoichiometry)(rate)
The two reactions shown in Figure 17 are mathematically equivalent. The representation in the first reaction in Figure 17 is the recommended practice, because processing applications are not required to be able to extract the stoichiometry from an explicit MathML definition such as the one shown in the second reaction.
Explicit mathematics should only be used in cases where the implicit formulation would be inappropriate. Some examples of such cases are:
-
If the stoichiometry of a reaction is unknown, but the modeller still wishes to relate the rate of change of a particular chemical species to the general reaction rate. Defining the
stoichiometryattribute implies that the stoichiometry is known to equal the value of that attribute. - If the modeller wishes to experiment with the stoichiometry of a species in different simulations using the model. (In this case, it might be easier if the stoichiometry is defined as a variable.)
- If the math implied from the recommended formulation would be incorrect, i.e., in the rare cases when a more complex function is needed to relate the change in concentration of a species to the reaction rate.
In all of these cases, it is recommended practice to put the mathematical expression used to define the change in concentration of a species inside the <role> element contained in the <variable_ref> element referring to the variable representing the concentration of that species.
Figure 17
The top <reaction> element shows the recommended method for defining the change in concentration delta_A of a chemical species A with respect to the reaction rate r. The second <reaction> element shows an equivalent representation using an explicit MathML definition. Use of this formulation is not recommended. The MathML blocks defining the rate laws are omitted.
It is an error to explicitly declare mathematics that conflicts with or duplicates implied mathematics. Therefore, a modeller cannot declare a stoichiometry attribute and delta_variable attribute in addition to explicit math relating the change in concentration of the referenced species to the reaction rate.
7.5.4 Meaning of mathematics in reactions
Equations defined in <math> elements in the MathML namespace inside a <role> element must be relevant to the the variable referenced by the parent <variable_ref> element, acting in the role defined by the value of the role attribute. This means that:
-
If the
roleattribute value is"rate", the equations must calculate the kinetic rate law (i.e., calculate the value of the referenced variable). Intermediate calculations related to the calculation of the rate are also allowed. -
If the
roleattribute value is"reactant"or"product", the equations must calculate the relationship between the general reaction rate and the rate of change of the species represented by the referenced variable (i.e., calculate the value of variable named in thedelta_variableattribute). Intermediate calculations related to the calculation of the delta variable are also allowed. -
In all other cases, the equations must relate an intermediate variable used in the rate calculation to the variable referenced by the containing
<variable_ref>element. For example, it would be appropriate to calculate an effective concentration of an inhibitor or catalyst in the<role>element contained in the<variable_ref>element that references the variable representing the actual concentration of that species.
7.5.5 Resolution of inconsistencies
Duplication of information is avoided as much as possible. However, because modellers must be free to define arbitrary rate laws, it was not possible to eliminate all information duplication. For instance, we cannot expect software to be able to deduce all information about a reaction from kinetic laws of arbitrary form, even though most information is in fact represented in these laws. Therefore, there is a possibility that the information in the mathematics and the information in the <reaction> element may be inconsistent.
It is anticipated that most modellers will define CellML models using some sort of processing software, which can reasonably be expected to write consistent CellML. However, since CellML is a text-based format, modellers may also create or edit models by hand, and in doing so risk creating inconsistent models.
The following rules govern the required behavior of CellML-compliant processing software in the event that information in the mathematics and the information in the reaction element do not agree:
- Preference is given to mathematics explicitly defined using MathML when running a simulation with the model.
- CellML processing software is free to determine which information to use in representing the model. Software is free to ignore the mathematics when creating a pathway diagram or chemical expression rendering of the model. However, software should clearly document which information is used to create representations of the model.
-
Processing software may check for inconsistencies between the mathematics and the information in the
<reaction>element. However, it is not required to do so, and it is left to the processing software to determine what to do if an inconsistency is found.
8 Metadata Framework
8.1 Introduction
Metadata is "data about data". In a CellML document, the principal data defined is the structure and mathematics of a biological model. Information that provides context for this data is metadata. Metadata can be included in a CellML document to facilitate searches of collections of models and model components. It provides a means for a modeller to include structured descriptive information about the model, which can help other modellers determine whether they can incorporate the model into their own work.
The CellML metadata structure is defined in a parallel document. This section of the CellML specification presents a framework for the use of metadata in a CellML document.
8.2 Basic Structure
Metadata is defined in a CellML document using the Resource Description Framework (RDF), which is a W3C recommendation. Two CellML RDF Schema are being developed for the convenience of model authors and developers of CellML processing software. The first schema will define a data model for storing elements from the Dublin Core element set, modification history information, inline documentation and specific biological metadata. The second schema will define how information about literature references should be stored in a CellML document. This schema will be an RDF serialization of the Object Management Group's Bibliographic Query Service (BQS) data model. The CellML RDF Schema will be defined and discussed in a companion metadata specification.
The table in Section 2.2.2 defines five metadata namespaces that CellML processing software is expected to recognise, and recommended prefixes to which these namespaces should be mapped. RDF elements are placed in the RDF namespace, which should be mapped to the prefix rdf. Dublin Core elements and Dublin Core qualifier attributes are placed in the appropriate namespaces, which should be mapped to the prefixes dc and dcq, respectively. CellML metadata elements and BQS citation elements each have their own namespace, mapped to prefixes of cmeta and bqs, respectively.
CellML processing software is free to ignore any and all metadata. However, it is hoped that software will at least display metadata. Model authors are free to develop their own RDF schema for metadata, or to store metadata in another format by using the CellML extension mechanism described in Section 2.2.3. However, doing so decreases the likelihood that CellML processing software will be able to do anything useful with the metadata in the model.
Metadata is defined within an <RDF> element in the RDF namespace, as shown in Figure 18. The recommended practice is to define the RDF namespace and any namespaces used by the enclosed metadata on the RDF element, even if these namespaces are already defined on the <model> element. This increases the re-usability of the RDF block. Furthermore, RDF processing software that does not recognise the CellML namespace can still parse a CellML document, extract the RDF blocks, and perhaps provide useful functionality with the information described in the RDF.
The <rdf:RDF> element contains an <rdf:Description> element, which defines an about attribute. The value of the about attribute must be a valid Uniform Resource Identifier (URI). A URI that points to a resource in the current document consists of a hash (#) followed by the value of that resource's id attribute.
Metadata is associated with a CellML document by assigning the about attribute an empty value (""). Any CellML element that has associated metadata must define an id attribute in the CellML metadata namespace (defined in Section 2.2.2). This attribute is of type ID, as defined in the XML specification. Its value must be unique across the CellML document, but need not have any meaning. Metadata is associated with a CellML element by assigning the about attribute on the <rdf:Description> element a value equal to the value of the cmeta:id attribute on the CellML element.
An RDF block should be stored in the element about which it contains metadata. This makes the element more re-useable. Elements in the MathML namespace are an exception to this recommendation. The MathML content of a <component> element might be extracted for use in a general MathML processor, which might not be able to handle RDF content. Therefore, metadata on MathML elements should be placed in the containing <component> element. If the RDF block contains metadata about the CellML document, it should be included in the root element of the document. Note that simply putting the RDF block inside an element is not sufficient to indicate that the metadata in the block refers to that element. The about attribute on the <rdf:Description> element must be used to indicate about which resource the RDF block contains metadata.
8.3 Examples
Figure 18 demonstrates the use of metadata in CellML. Three RDF blocks are shown: one that provides metadata about the CellML document, one that provides metadata about the model, and one that provides metadata about a component contained in the model. Only the RDF framework elements are shown. The actual metadata is not shown here. Examples in the companion CellML metadata specification will demonstrate how to use the recommended metadata elements.
Figure 18 An example of the use of metadata in a CellML document.
The first RDF block provides metadata about the CellML document. This is indicated by the empty value of the about attribute on the <rdf:Description> element. The second RDF block has a value of "#model01" for the about attribute on the <rdf:Description> element. This indicates that this metadata provides information about the model that is delimited by the <model> element with an cmeta:id attribute with a value of "model01". The final RDF block provides metadata about the membrane component. This is indicated by assigning a value of "#comp01" to the about attribute on the <rdf:Description> element.
Note that all three RDF blocks declare the RDF and CellML metadata namespaces. This makes the RDF blocks portable: the information needed to interpret the RDF will be preserved even if the blocks are extracted from the CellML document.
8.4 Rules for CellML Documents
8.4.1 The <rdf:RDF> element
- Allowed use of the
<rdf:RDF>element-
Any CellML element may contain any number of
<rdf:RDF>elements. [ Metadata may appear on any CellML element, and may be split across multiple<rdf:RDF>elements. The recommended practice is to enclose all metadata about a particular element in a single<rdf:RDF>element. In this and subsequent rules, the use of therdfprefix indicates that elements and attributes are in the RDF namespace. ] -
The content of an
<rdf:RDF>element must conform to the Resource Description Framework (RDF) Model and Syntax Specification recommendation from the W3C. [ Avoid the abbreviated syntax defined in the recommendation to ensure maximum portability of the metadata. ]
-
Any CellML element may contain any number of
8.4.2 The <rdf:Description> element
- Allowed use of the
<rdf:Description>element-
The content of an
<rdf:Description>element must conform to the Resource Description Framework (RDF) Model and Syntax Specification recommendation from the W3C. [ The recommended practice is for contained elements to adhere to an RDF schema, and to use the CellML metadata schema wherever possible. ]
-
The content of an
- Allowed values of the
aboutattribute-
The
aboutattribute on an<rdf:Description>element must either be empty or have a value equal to a valid URI that points to an element in the current document (i.e., is equal to the value of acmeta:idattribute on an element in the current document preceded by a hash (#)). [ An<rdf:Description>element with an emptyaboutattribute contains information about the CellML document. An<rdf:Description>element with anaboutattribute that references acmeta:idattribute value contains information about the element in the current document identified by thecmeta:idattribute. ]
-
The
8.4.3 Proper use of the cmeta:id attribute
-
The
cmeta:idattribute may appear on any element in a CellML document. [ In this and subsequent rules, thecmetaprefix places elements and attributes in the CellML metadata namespace. ] -
The value of the
cmeta:idattribute must be unique across the CellML document. -
A
cmeta:idattribute must be defined on any element in the CellML or MathML namespaces for which RDF metadata is defined.
8.5 Rules for Processor Behaviour
8.5.1 Metadata is optional
All metadata is optional. A model without any metadata is a valid CellML model. However, we strongly recommend that the modeller provide as much metadata as possible, particularly his/her name and contact information and a reference for a paper that describes the development of the model.
8.5.2 Associating metadata with resources
Software must associate the metadata contained within an RDF block with a CellML document, a CellML model, or a specific element within the CellML model according to the following rules:
-
If the
aboutattribute on an<rdf:Description>is empty, then the metadata contained within the<rdf:Description>element refers to the entire CellML document. -
If the
aboutattribute on an<rdf:Description>points to a<model>element, then the metadata contained within the<rdf:Description>element is associated with the referenced model. -
If the
aboutattribute on an<rdf:Description>points to any other element within the current document, then the metadata contained within the<rdf:Description>element is associated with the referenced element.
8.5.3 General meaning of metadata
Metadata may refer to the CellML document, the CellML model, or a specific element within the CellML model. The following list documents the intended meaning of metadata on each of these resources. More detailed information can be found in the companion CellML metadata specification.
- Metadata that refers to the CellML document provides information relevant to the document as a whole, independent from the use of the document to specify a model. Examples of metadata that might appear on a CellML document are last modified date (date on which the document was last edited) and publisher (person or organization distributing the document).
- Metadata that refers to the CellML model provides information relevant to the model as a whole. For instance, the model author is the person who created the complete model, even if some of the components were taken from a shared database and have different authors.
- Metadata that refers to a specific CellML element provides information about that element only. It does not provide information about elements that are contained in the referenced element.
| |

