The (now defunct) reaction element in CellML
Contents
- 1. What is the Reaction Element?
- 2. Removing the Reaction Element
- 3. Re-writing the Reaction Models
- 4. Archiving CellML Models Containing the Deprecated Reaction Element
1. What is the Reaction Element?
The flexibility of the CellML language means it can be used to describe many different physiological processes, occurring over a wide range of spatial and temporal scales. Such processes include signal transduction pathways, metabolic pathways, cell division cycles, and electrophysiological phenomena in excitable cells. While it is possible to specify a model purely in terms of equations expressed in MathML, when the CellML language was first defined a reaction element was included. This reaction element was designed to describe individual steps in a reaction pathway or network, and it included a description of the reaction kinetics, the identities of the reactants and products, and it also identified any reaction catalysts (such as enzymes) or inhibitors. The inclusion of the reaction element in the CellML specification was justified because it was felt valuable information about the reaction would be lost if the model was reduced to a purely mathematical description.
To illustrate the reaction element, imagine an irreversible reaction where two reactants (A and B) form a single product (D). This reaction is catalysed by an enzyme (C), and it is inhibited by a negative feedback loop by the product D.
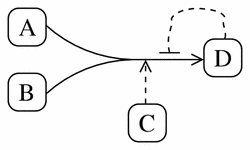
Using the reaction element in CellML this would appear as:
<reaction reversible="no">
<variable_ref variable="A">
<role role="reactant" delta_variable="delta_A" stoichiometry="1" />
</variable_ref>
<variable_ref variable="B">
<role role="reactant" delta_variable="delta_B" stoichiometry="1" />
</variable_ref>
<variable_ref variable="C">
<role role="catalyst" />
</variable_ref>
<variable_ref variable="D">
<role role="product" delta_variable="delta_D" stoichiometry="1" />
<role role="inhibitor" stoichiometry="1" />
</variable_ref>
(For a more detailed explanation of the reaction element please refer to the appropriate section of the CellML 1.1 specification).
2. Removing the Reaction Element
In practice we found implementing the reaction element in a CellML model often required the equations to be re-written, such that they no longer reflected those in the original publication. At best this created extra work and some confusion, at worst it broke the model. Further, it was not in keeping with the general philosophy that the CellML model itself should reflect the pure, published mathematics. Consequently the decision was made to remove the reaction element from the CellML 1.2 specification, and instead of storing reaction data in the model itself it will, in future, be stored in the metadata associated with the CellML model. In particular, we intend to implement the use of an ontology to label the model variables as reactants, enzymes, products etc.
3. Re-writing the Reaction Models
In the meantime, we were left with many models in the CellML repository which included the reaction element. The majority of these have since been re-written and the reaction element has been replaced by simple mathematical equations.
In most cases we found that the structure of a model written with and without the reaction element was so different, it necessitated a complete re-write of the model. In this way we were able to translate approximately three quarters of the reaction models in the repository. Where we were unable to translate the models it was usually because the original published paper did not contain sufficient information. In particular the reaction element supported qualitative models, defined as models which consist solely of information about how the different chemical species in the pathway relate, and they contain no mathematics. As such no mathematical representation of these models exist.
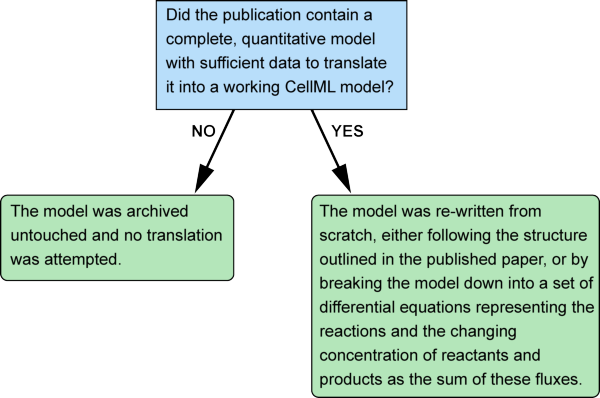
In practice there were just two steps in the process of removing the reaction element from a model:
4. Archiving CellML Models Containing the Deprecated Reaction Element
Models containing the reaction element were identified of the majority were successfully re-written as outlined above. However, at the end of this translation process we were left with 13 models which we were unable to re-write. The main reason for this is because the CellML models were originally taken from papers (often review papers) which did not contain a (complete) mathematical model. These qualitative models were usually based on a schematic figure of a reaction pathway. All the reaction kinetics were assumed to be mass action and all the parameters were assigned a default value of 1.
Although it is possible to recode these models to remove the reaction element, the end product will be pretty meaningless, and the CellML model will not represent a published model. Consequently we have removed these 13 models from the main repository and they are now listed below as a legacy of a learning process.
haugh_wells_lauffenburger_2000
hefti_harder_eppenberger_schaub_1997
kodama_fukuda_pan_sano_takahashi_kato_makino_manabe_murata_ogawa_2000

