CellML Tools Plan
Note
This is document is a draft attempt to plan CellML Tools in more detail. The contents of this document have been approved by the CellML team. However, it is only a rough roadmap, intended as a way to help determine a rough list of what modules and steps are likely to be required, to guide development of the final project. For a more general overview, see the planning document at http://www.cellml.org/tools/mozCellML/Planning .
Overall architecture
- The architecture is split into three parts: Infrastructure, Tools, and Services. Tools and services are collectively called modules.
- Infrastructure provides an environment in which tools and services operate.
- Tools provide a user-interface, but perform little substantive work (they just call through to services).
- Services perform backend tasks, for example, integrating a CellML model.
- All tools developed in the institute use the Mozilla framework, using XUL and Javascript.
- The CellML Context (see below) provides part of the infrastructure to link tools and services together.
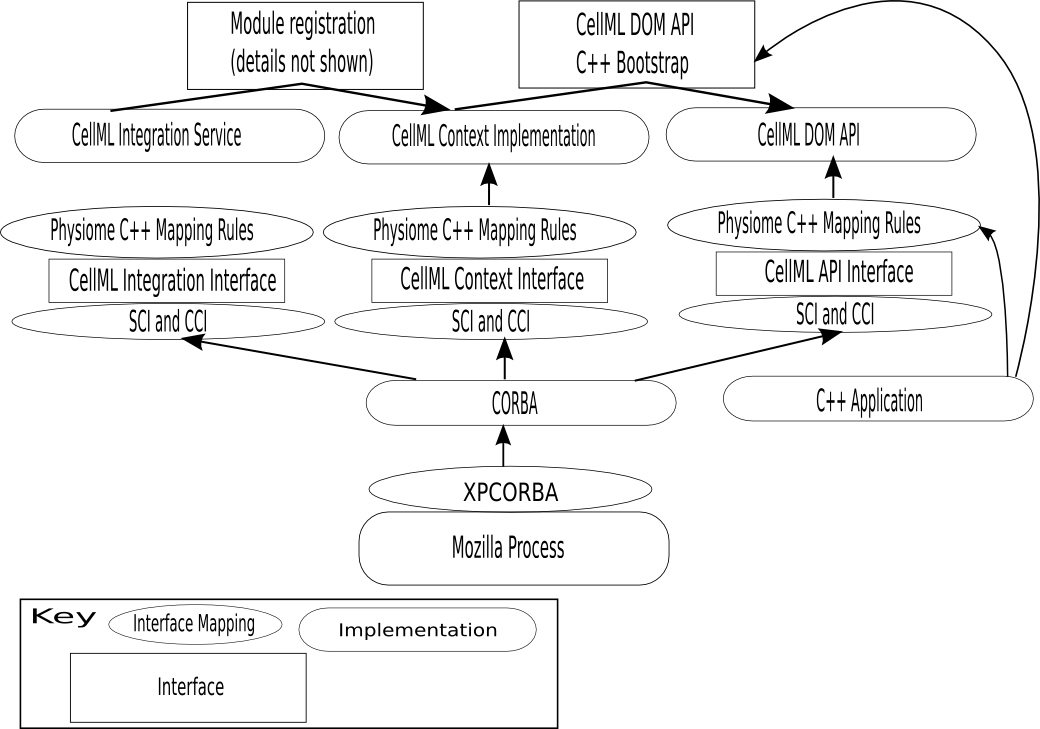
Interface specifications
- Communication between modules only occurs across well-defined interfaces.
- Interfaces are specified in OMG IDL, and should have comments in them further describing what they do.
- Interfaces are not tied to any particular implementation, so it is possible to switch between implementations which support the same interface, providing flexibility and allowing Inversion of Control based designs.
Physiome C++ Mapping
- The Physiome C++ Mapping (PCM, formerly the Simple C++ Interface) can be generated from OMG IDL files, using a compiler. These can be used both to implement interfaces, and to call them (therefore, there is no middleware between the call and the implementation). Such code can work because all methods on the generated base header are pure virtuals.
- CCI (CORBA Calling Interface, an implementation of a simple C++ interface which forwards calls to a CORBA object reference) and SCI (Simple Calling Interface, a CORBA servant which forwards calls to an object implementing the Physiome C++ Mapping. The 'simple' in the name exists for historical reasons) can be generated from the OMG IDL. This allows calls between CORBA and code using only the physiome C++ mapping.
- The interface generation, SCI generation, and CCI generation have already been developed.
- The Subversion repository in which this code is stored is publicly readable at https://svn.physiomeproject.org/svn/physiome/simple_interface_generators/trunk/
The CellML API
- The CellML API provides a representation of CellML models, and allows for models to be read into memory.
- It is written using the Physiome C++ Mapping.
- It allows for annotations of any element in the CellML model.
- It allows events to be set on changes to the model.
- The CellML API, including events, and a testsuite, have already been developed.
- An older (pre-events) version is at ftp://ftp.bioeng.auckland.ac.nz/pub/physiome/cellml_api/cellml_api-0.1/CellML_DOM_API-0.1.tar.gz .
- The Subversion repository is publicly available at https://svn.physiomeproject.org/svn/physiome/CellML_DOM_API/trunk/
The CellML Context
- The CellML Context provides a central part of the infrastructure, through which all other modules can be accessed. The CellML Context also provides services for managing which CellML models are currently loaded, and storing version histories of those models, and provides access to the CellML API.
- The CellML Context allows annotations of arbitrary classes. This allows for special handlers for each type of object to be installed, allowing for extensible user interfaces to be designed.
- The CellML Context is written in C++ using the simple C++ interface. It could theoretically be accessed directly from C++, but will mainly be used over CORBA, so that it will work across processes.
- The CellML Context implementation has already been developed.
- The CellML Context is shipped as an optional module of the CellML API, in the directory sources/cellml_context.
The CellML CORBA Server
- The CellML CORBA Server provides access to the CellML Context across CORBA. It saves the IOR of the CellML Context at a well-known location in the user's home directory (i.e. for a given platform, the rules for determining the save location will be the same). Services and tools read this file and connect to the Context on that IOR.
- The CellML CORBA server has already been developed.
- The CellML CORBA Server is shipped as an optional module of the CellML API, in the directory sources/cellml_corba_server.
xpcellml_api
- xpcellml_api is a Firefox extension (which could also be made to work with, e.g. XULRunner) provides access to the CellML Context in the Mozilla environment, by using a derivative of XPCORBA.
- xpcellml_api has already been developed.
- The xpcellml_api Subversion tree is available at https://svn.physiomeproject.org/svn/physiome/xpcellml_api/trunk/
The Physiome CellML Environment(PCEnv)
- The Physiome CellML Environment runs in the Mozilla environment, and is written in XUL and Javascript, but communicates via xpcellml_api to make use of the CellML API and various CellML services (all of which it locates using the CellML Context).
- PCEnv will follow an interface which allows extensions to be added.
- PCEnv allows both editing of models, and numerical integration of models of various kinds.
- It allows numerical integration of models of various kinds.
- It allows graphs of the models to be drawn and manipulated on a canvas element.
- As this tool may process large amounts of data, certain parts of the UI code may need to be written in C++.
- There is currently a Java program which performs the editing task, although most code will need to be re-written to work together with the CellML API and context.
- A version of PCEnv which allows graphing and integration has been developed. It does not yet support full editing capabilities, but this is planned for after the first release.
- Regular snapshot releases of PCEnv binaries are available at https://svn.physiomeproject.org/svn/physiome/snapshots/pcenv/trunk/ . Go into the directory for your platform and see index.html for a list of snapshot builds.
- The Subversion repository can be accessed at https://svn.physiomeproject.org/svn/physiome/pce/trunk/
The CellML (C?) Code Generation Service(CCGS)
- The CCGS takes a CellML model and produces code of a certain kind.
- The CCGS is implements a Physiome C++ Mapping of the CCGS.
- CCGS support is now working.
- It is an optional extension shipped with the CellML API, in the CCGS subdirectory of the source tree.
The CellML Integration Service(CIS)
- The CIS allows models to be numerically integrated, and the results obtained.
- The initial implementation of the CIS will include a complete gcc distribution, and will use the CCGS to generate code. The resulting program will then run, and communicate back to the CIS using a specialised protocol.
- CIS support is now working.
- It is an optional extension shipped with the CellML API, in the CIS subdirectory of the source tree.

