CellMLModelRepositories
This section is about the systems related to CellML repositories.
CellML Model Repository
See also CellMLRepositoryMigration for a discussion on how we are setting out to implement this, and CellMLRepositoryWorkflow for a discussion on the workflow governing model curation, review, and publication.
Objectives
- Promote the sharing of models
- Accessible to the public
- Provide useful software level interfaces for accessing, querying, and storing models.
- Provide a curatorial work-flow where models can be submitted, reviewed, validated, and published.
- Provide a mechanism for people in various roles to provide feedback on models.
- Provide a mechanism for advertising new models or changes to existing models.
Operation
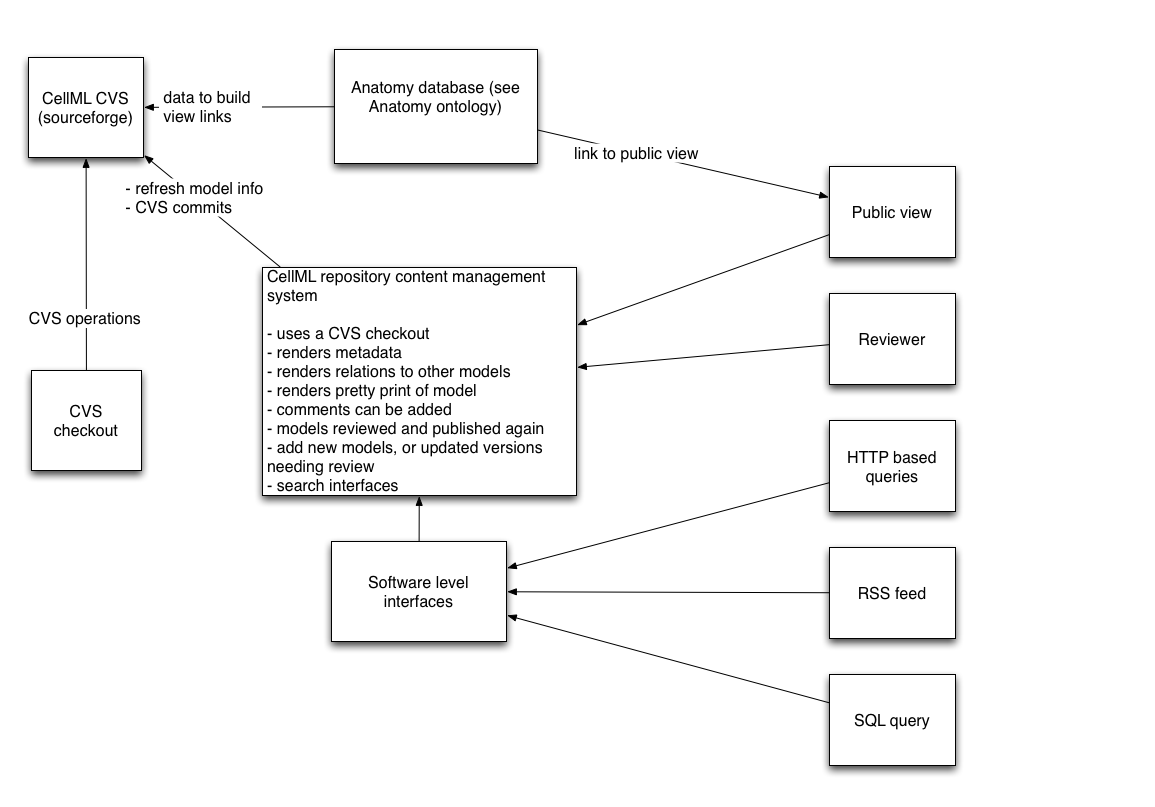
A diagram of the system is shown in Repository Design
How does the repository work?
CVS subsystem:
- A CVS repository is the primary source. Users can make public checkouts of it for local use. Regular releases of the repository in archive formats could be made available.
- People with developer access are allowed to modify the CVS repository directly.
- Any model validation checks are run on regular automated checkouts of the repository, and the appropriate people notified by email.
- The version system that CVS uses and the version system that is relevant to the models (CellML model version) themselves will be independent from each other. The CellML model version will be added as a CVS tag, so that older versions can be located and served.
Content management subsystem:
The content management system provides a human and software level interface to the repository.
Public viewers are able to:
- review the meta-data and pretty print of a model
- download individual models
- add comments; for example, add comments, suggestions for corrections or improvements
- submit new or changed models for review
Reviewers run validation tests on models that are waiting review and submit them to the CVS, which also makes them published to the public.
Model developers are able to change, or add new models, which then become part of the CVS repository once they have passed some levels of validation.
Notification about new models or changes to existing ones will be made via a human readable interface, such as new items, or recent changes page, as well as through software level interfaces such as RSS feeds.
The software interfaces provide:
a range of interfaces for querying the repository and returning data sets. They also provide a mechanism for adding new models or updating others.
the main HTTP interface for all the models and all their versions. This will be the main HTTP layer interface for resolving Uris in imports relevant to the repository.
Sample URL for a particular version of a model:
http://www.cellml.org/cellmlmodels/2004_10_hunter_nielsen_1?&version=1.0
access at the software level to query interfaces using SQL or HTTP based query string.
software level submission of new or changed models. Depending on the status of the authenticated user, the models may or may not need to pass review.
While RDF and OWL based queries are an ultimate goal, it is perhaps better to provide at first a SQL interface. This is familiar to many more developers and still provides a good mechanism for doing some cross result set Boolean operations.
Naming a model in a repository should follow a best practice guide. See CellML Model Naming for a discussion of this and a proposal.
Design of the repository system
Implementation
The CVS repository would reside on sourceforge. Developers would need to create member logins and an administrator can add them as repository developers.
The CellML repository content management system would be implemented as a Plone site [1]. The model objects will be archetype wrappers around python object representation of CellML models. There are a few work-flows that are available to plug-in [2], or customized ones could be added.
SQL interfaces would be implemented using Zope based SQL interfaces to the CellML model objects stored in the repository.
RSS feeds would be implemented using the Zope syndication capability, perhaps modified to enable search based RSS, which are RSS feeds based on a specific Plone search query.
HTTP based queries would be translated into a Plone search, or SQL query, or method calls on a custom python based query object.
| [1] | Plone Site |
| [2] | Plone Workflows |
CellML Model Naming
Considerations
- human readable names do help some understanding of what is in a model.
- there should be a set of best practices
- a name should always remain stable, a change in name is considered to be a variant of the original model. See CellML Model Versioning.
- what is considered to be a version update should not force a change in name; therefore, versions should not be included in the name. See CellML Model Versioning
- it is difficult to place a model exactly in one biological category, so using these to help name a model should be avoided.
- the meta-data of models should be readily available to users and editors so that names are not used to hold too much meaning
Useful references:
Formula for naming CellML models
The model name formula is:
$modelauthor1_$modelauthor2_..$modelauthorN +publication year +a,b,c... (optional) +_versionNN +_variantNN (optional) +_partNN (optional)
$modelauthor1_$modelauthor2_..$modelauthorN are the last names of authors in publication order.
publication year is the year of publication of the article defining the model.
a,b,c... resolves conflicts with the same author list publishing more than one model in a calendar year.
partNN serves to differentiate the constituant components of a CellML 1.1 model.
The URI for model import and metadata is built using:
physical location+/name.suffix
where location and suffix are implicit. Models will be stored at the http://www.cellml.org/cellmlmodels/ address.
Example: this model which is currently called:
http://www.cellml.org/examples/models/glycolysis_pathway_1997.xml
would be called:
http://www.cellml.org/cellmlmodels/rizzi_baltes_theobald_reuss_2004_version01
The form of authors has some problems:
- author order. It would be nice to apply a simple rule such as alphabetical, and leave arguments about order of authorship to the meta-data.
- encoding - characters outside of ASCII need to be mapped into ASCII. We cannot use URL encoding since we want to be able to map these URIs to file system paths, as well as the reverse, i.e. to be able to engineer the URI from the file-system.
- the current formula uses underscore '_' as a delimiter. I did consider sub-folders, but there is really no argument to have them. An important consideration is that files may be taken out of their original context, so lose some meaningful information if they were deeply nested in folders.
CellML Model Versioning
Reasons to change a model
Fix errors
Errors in models are fixed for the reasons:
- so that it validates as a CellML model
- to fix math
- so that it is a true representation of the publication, i.e.
- to match the architecture of the published model - i.e. the abstractions they make
- to make the mathematic formulations the same
- to generate the same results from simulations; however:
- there may be differences in simulation systems, which is outside the scope of the CellML model
- there results may be incorrect w.r.t the published model
- there may be an update/amendment/errata to the publication that correct errors in the original one
Change model
A model may be changed for the reasons:
- a different formulation of the math - CellML models that use reactions perhaps have some examples of these - they may not exactly represent the equations of the model. Another candidate is changing some math so that the model can run as a simulation, for example, to break cycles that are in the published math, but which are expected to be modified in simulations.
- new structures, math, or relationships are required to achieve the results that were published with the original model
- general evolution of models
- to promote reuse - e.g. break down the model into smaller models that are imported. While the overall model should be the same semantically, the syntactical representation has changed.
- add new meta-data. But this should not impact the versioning of the model itself, though it is usually associated with a change that would. Meta-data versioning should be an ontology issue.
Method for determining a variant vs version
A simple method for determining a new version or a new variant could be :
- if an error is fixed, then it is a new version. An exception to this is errata - where we should be able to make a variant that implements this errata and which is actively promoted as the more correct primary version of the model.
- if a change is made, then it is a new variant. There is an argument that a change to make use of imports does not change the overall semantic structures, and therefore will not break other models that import components from it. This may only require a version change. However, if we are changing a CellML 1.0 to a CellML 1.1 type model, then perhaps it is a variant.
How do we name these versions and variants?
Versions do not change the model name; therefore, we need to add another structure to CellML core, such as a version element. We could add this in the meta-data - but it would be nice not to have to use RDF to get at it.
Variants are more difficult. A new variant may be a very big step that someone else wants to take ownership of and publish. It should have a new name, and so should be accommodated by the naming practices.

