Translating a mathematical model into CellML
About this Tutorial
The creation of a CellML model is a multi-step process which starts with the identification of a model in a published paper, followed by its translation into CellML, simulation, and curation, and finally, ends with the model being uploaded into the CellML Model Repository. Having said that, the process doesn’t really end here either. The models in the repository are free for download, and can be further modified and subsequently re-submitted as a new version.
This document is intended to discuss, in slightly more detail, the multi-step process we undertake to create a CellML model.
Identification
A mathematical model of interest is identified for translation into CellML. The usual source of such models is peer reviewed publications.
CellML can be used to express models describing a wide range of physiological processes, including metabolism, electrophysiology, signalling, cell division, immunology, muscle contraction, etc.
The suitability of a published model for translation into CellML is assessed by criteria such as:
- Does the paper contain a complete – or near complete – mathematical description of the model?
- Does the paper include a full set of parameter values?
- Have unit definitions been considered?
- Is the model within the scope of CellML? For example, models containing spatial elements and/or time delays are currently not supported.
Creation
The published model is translated into CellML using the editing tools OpenCell, Cellular Open Resource (COR) and JSim, and/or a simple text editor such as Notepad++.
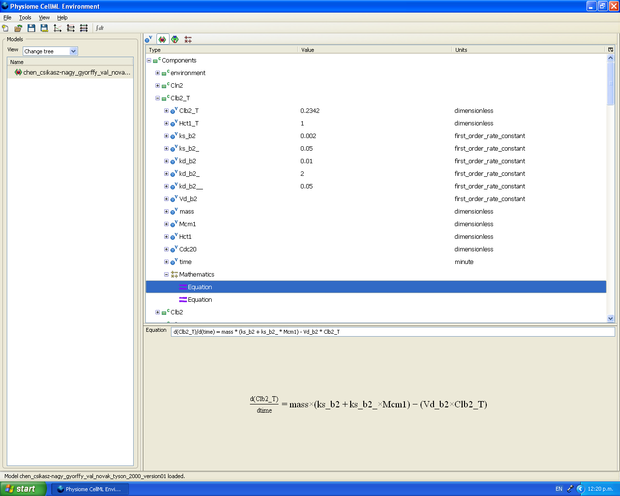
Editing a model in OpenCell
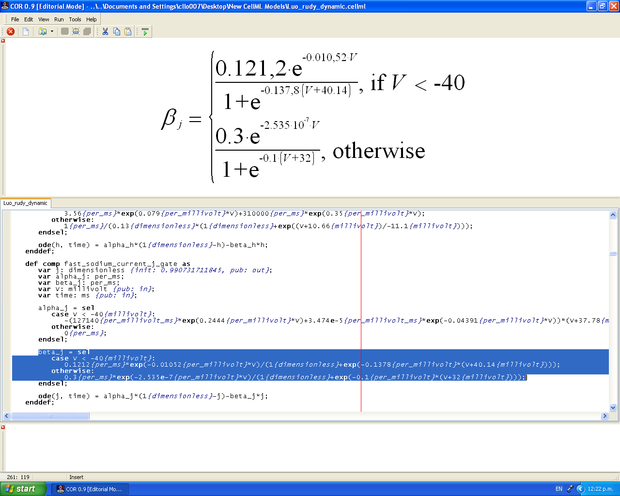
Editing a model in COR
Mathematical equations are expressed in MathML, and metadata are expressed using Resource Description Framework (RDF) and the Web Ontology Language (OWL). Metadata may include the identity of the model author, details of the original publication, and biological data such as species, cell type, molecular IDs, etc. Currently the tools OpenCell and COR will support the presence of metadata in a model, however there are no facilities for annotating models with this type of information. Therefore this kind of data has to be added to a model by hand in a text editor. Alternatively, citation information, modification history, and cutation status metadata can also be added to a model via a web interface when uploading a model into the repository (see section 7 below for more information).
Simulation
Once the translation into CellML is complete, the model is loaded into the tools OpenCell and/or COR and is simulated. Simulation parameters are defined with the intention of recreating the original results from the published paper.
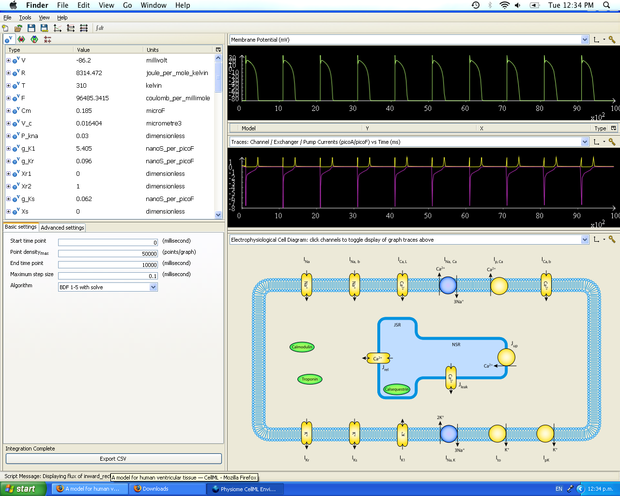
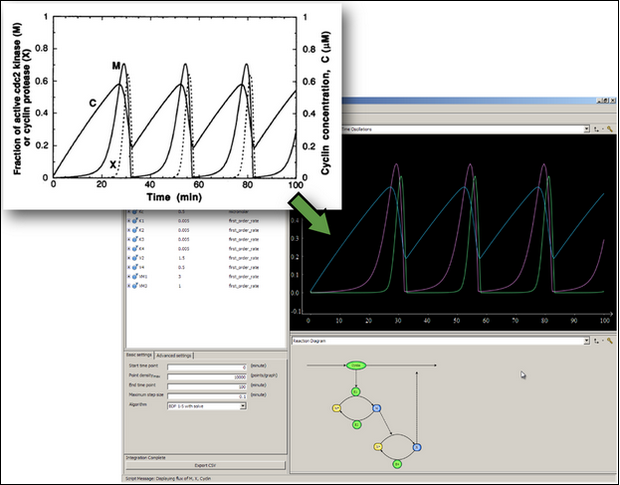
Simulation in OpenCell
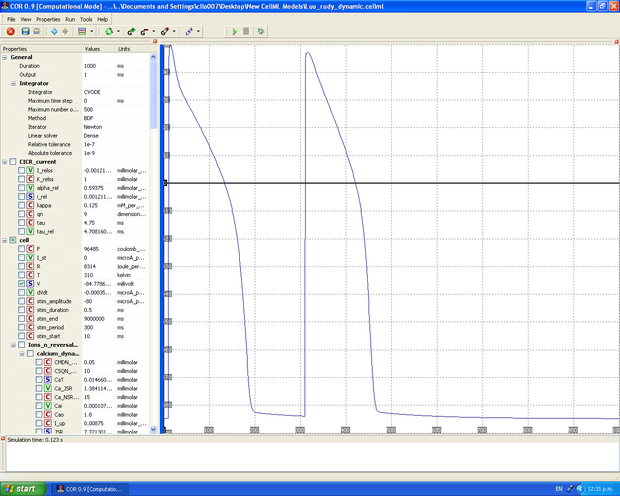
Simulation in COR
The ultimate aim is to get the graphical output from the CellML model to match the graphical simulation figures in the published paper:
Curation
It is rare for a CellML model to reproduce the published results at the first iteration.
Type errors in the paper, or missing parameters, equations or unit definitions, mean the CellML model is often incomplete, contains errors, or unit inconsistencies. At this point we seek help from the original model author and, where possible, obtain a copy of their original code.
A ‘star' system signifies the curation status of a CellML model:

Of the approximately 360 models in the model repository, more than half have been curated to some degree (one star or more), and a third have two stars, implying they recreate the published results in the original paper. For more details on curation and the star system please refer to the document on model curation.
Documentation
For each model, we endeavour to produce an abstract-style description, together with links to the original publication, and a schematic diagram. Further, where possible, we create a OpenCell session file, which includes an embedded SVG diagram to facilitate navigation of the model.
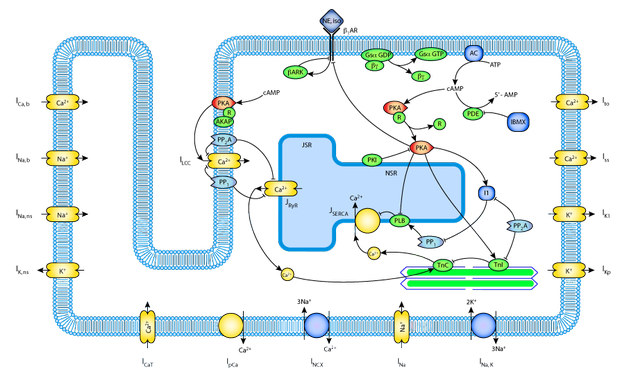
Schematic diagram for the Saucerman-McCulloch 2004 cardiac myocyte model
Repository
Once the model is complete it is loaded into the model repository.
A snapshot of the model list in the repository.
Once it is loaded into the repository, the model is available for free download. It can be used, potentially updated, and a new version can be uploaded into the repository.

