Modularity in CellML 1.1 tutorial
Introduction to CellML 1.1
Making CellML models available to the public, in an open source repository, is an important part of model communication, but it does not necessarily lead to easy model reuse. One of the advantages of using CellML1.1 over CellML 1.0 as a modelling language is the "import" feature, which allows existing models to be connected together; for example, individual pathways may be connected together to form large networks. Further, this feature also facilitates model reuse amongst researchers, and it also allows researchers to build upon previously published models, without the need to start from scratch.
Currently, the vast majority of the models in the CellML Model Repository are in the CellML 1.0 format. This is, at least in part, a legacy of the process of language evolution; CellML 1.0 was developed first and was adopted from the onset of the project, while CellML 1.1 is a more recent development. In addition, the tools have only relatively recently been able to edit and simulate CellML 1.1 models, whilst the software behind the model repository has yet to support them (although this will be resolved with the implementation of the new repository in April this year).
It is our long term goal to use CellML 1.1 to build a library of reusable biological model components, such as ion channels, pumps, exchangers, enzymes, substrates, etc., allowing the creation of new, more complex models from pre-existing parts.
The language
CellML 1.1 has two key features which make it applicable to model exchange and reuse:
- A modular architecture allowing models to be easily broken down into components; and
- The "import" feature facilitating model connectivity and reuse.
Two common examples of where model import, modularity and connectivity are useful:
1) Models can evolve over time:
Often biological models are formed from similar components - or modules - and they can build on each other, becoming increasingly complex over time. Take for example the historical sequence of cardiac ventricular cell models. Numerous models have been published, building on each other and increasing in complexity, over a period spanning several decades (Nickerson and Hunter, 2006).
Choosing just three of these models, the schematic figures below show how the models have increased in complexity:
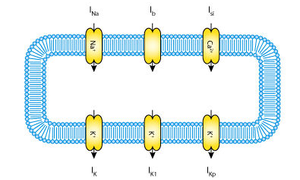
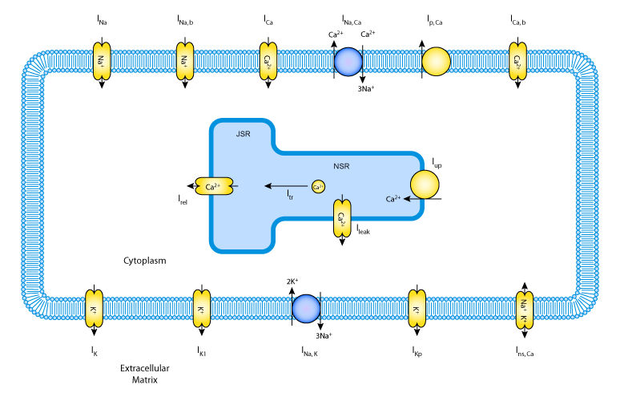
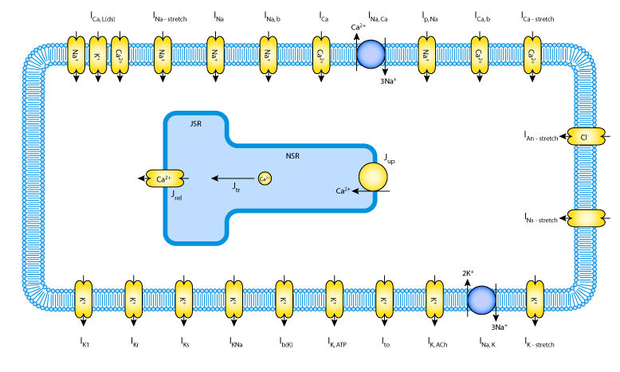
Rather than starting from scratch with the creation of each model, CellML 1.1 allows the modeler to import an earlier model, and then build upon it by adding new channels, pumps, and fluxes.
As mentioned above, eventually we would like to create a library of models each of which characterises a single channel, pump, ion, etc. and then connect these together, a bit like Lego bricks, to form a complex model.
Two papers (Nickerson et al. (2008) and Cooling et al. (2008)) have been published recently which expand on this idea, and demonstrate how CellML 1.1 can be used to build composite, complex models, from more simple parts.
2) Models can share entities and processes:
Some natural systems appear to have a modular composition, such as signalling and metabolic pathways. CellML 1.1 permits the reuse of components which describe shared entities (ligands, receptors, complexes) or processes (reactions) between models.
For example, consider these two metabolic pathways:
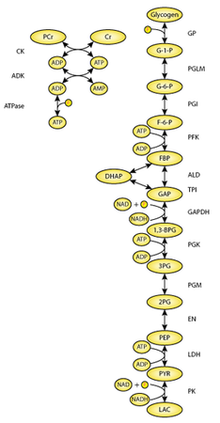
Glycolysis Pathway
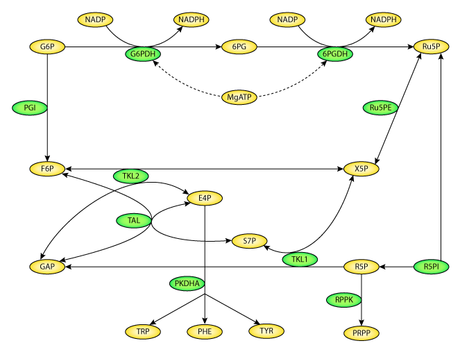
Pentose Phosphate Pathway
Several entities are shared, including G6P and F6P. Rather than start from scratch when writing these models, it would be ideal to reuse the G6P and F6P components.
For a more complex example of how CellML 1.1 can be used to build models which share entities and processes please refer to the recent publication by Terkildsen et al. (2008).
Implementing CellML 1.1
The CellML specification offers few guidelines for how to structure a model. A mathematical model of a biological process can be represented in CellML in many different ways, and the final structure of the model mainly depends on the individual author’s modelling style. While this flexibility is an advantage for the model developer, the resultant diversity in model structure can make it difficult to combine and reuse a particular biophysical concept.
A paper has been published (Wimalaratne et al. 2009) outlining a series of best practice steps to create a reusable CellML 1.1 model. We refer you to this publication for more details on how to create a CellML 1.1 model.
More recently, another paper has been published on the subject of modular modelling with CellML 1.1 (Cooling et al. 2010). This paper outlines the use of libraries of standard virtual biological parts to build up complex models.

